Biomarker and Prognostic Value of Super-ARMS Detection for EGFR Mutation in Advanced NSCLC
- PMID: 40655938
- PMCID: PMC12255349
- DOI: 10.2147/OTT.S518837
Biomarker and Prognostic Value of Super-ARMS Detection for EGFR Mutation in Advanced NSCLC
Abstract
Background: ctDNA is a non-invasive and convenient method for detecting EGFR mutations in non-small cell lung cancer (NSCLC). However, its sensitivity is lower than that of tissue-based testing. To enhance ctDNA detection efficiency, we identified the patient population most suitable for ctDNA testing, assessed the relationship between ctDNA and tumor markers, and examined the clinical significance of ctDNA in medical practice.
Methods: A single-center retrospective study was conducted, including 135 patients with NSCLC who underwent histological and liquid Super-ARMS tests. Of these, 92 patients with EGFR mutations detected in both tumor tissue and plasma were classified into the EGFRt+, p+ group, while 43 patients with EGFR mutations detected only in tumor tissue were classified into the EGFRt+, p- group. The clinical features and outcomes between these two groups were compared.
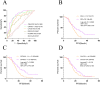
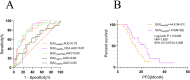
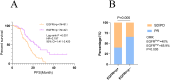
Results: The positivity rate of Super-ARMS test was 68.1% (92/135). The presence of EGFRt+, p+ in the Super-ARMS test was significantly associated with pleural effusion, bone, liver, and multiple organ metastases. Compared to the EGFRt+, p+ group, the EGFRt+, p- group had a significantly better PFS (P < 0.01). Carcinoembryonic antigen (CEA) levels demonstrated a strong predictive value for identifying plasma EGFR-mutated patients (AUC 0.828, sensitivity 68.8%, specificity 84.4%), while Maximum Standardized Uptake Value (SUVmax) also showed diagnostic value for plasma EGFR-mutated patients (AUC 0.78). Additionally, combination of TP53 and EGFR mutations in plasma provided improved risk stratification for PFS (P < 0.001).
Conclusion: Patients exhibiting metastasis, elevated levels of tumor markers and SUVmax are more suitable for plasma EGFR mutation testing in clinical NSCLC management. Moreover, a positive plasma ctDNA test not only guides targeted therapy but also predicts a worse prognosis.
Keywords: CEA; EGFR mutation; NSCLC; ctDNA; prognosis.
© 2025 Liu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Clinical implications of ctDNA for EGFR-TKIs as first-line treatment in NSCLC.J Cancer Res Clin Oncol. 2023 Mar;149(3):1211-1220. doi: 10.1007/s00432-022-03952-z. Epub 2022 Apr 5. J Cancer Res Clin Oncol. 2023. PMID: 35380256 Free PMC article.
-
First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer.Cochrane Database Syst Rev. 2016 May 25;(5):CD010383. doi: 10.1002/14651858.CD010383.pub2. Cochrane Database Syst Rev. 2016. Update in: Cochrane Database Syst Rev. 2021 Mar 18;3:CD010383. doi: 10.1002/14651858.CD010383.pub3. PMID: 27223332 Updated.
-
Prediction of the efficacy and clinical prognosis of first-line EGFR-tyrosine kinase inhibitors in non-small cell lung cancer patients based on ΔCt values derived from the super-amplification refractory mutation system (ARMS): a real-world retrospective study.J Thorac Dis. 2025 Jun 30;17(6):3897-3911. doi: 10.21037/jtd-2025-97. Epub 2025 Jun 25. J Thorac Dis. 2025. PMID: 40688317 Free PMC article.
-
Prognostic value of EGFR and KRAS in circulating tumor DNA in patients with advanced non-small cell lung cancer: a systematic review and meta-analysis.Oncotarget. 2017 May 16;8(20):33922-33932. doi: 10.18632/oncotarget.15412. Oncotarget. 2017. PMID: 28430611 Free PMC article.
References
-
- Subbiah V, Cote GJ. Advances in targeting RET-dependent cancers. Cancer Discov. 2020;10(4):498–505. doi: 10.1158/2159-8290.CD-19-1116 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

