Gene regulation in Cryptosporidium: New insights and unanswered questions
- PMID: 40656160
- PMCID: PMC12246861
- DOI: 10.1016/j.crpvbd.2025.100280
Gene regulation in Cryptosporidium: New insights and unanswered questions
Abstract
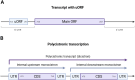
Parasites of the genus Cryptosporidium have evolved to have a highly compact genome of ∼9.1 Mb. The mechanisms that regulate gene expression in Cryptosporidium spp. remain incompletely understood at all levels, including chromatin accessibility, transcription factor activation and repression and RNA processing. This review discusses possible mechanisms of gene regulation in Cryptosporidium spp., including histone modifications, cis regulatory elements, transcription factors and non-coding RNAs. Cryptosporidium spp. are among the most basal branching apicomplexans and existing evidence suggests that they diverge from other members of their phylum via retention of the E2F/DP1 transcription factor family, and the recent discovery that C. parvum produces polycistronic transcripts. Most of what we know about gene regulation in the genus Cryptosporidium is based on sequence conservation and homology with other members of the phylum Apicomplexa, and in some cases, more distant eukaryotes. Very few putative gene regulatory components identified in Cryptosporidium spp. are supported by experimental confirmation. This review summarizes what we know about gene regulation in Cryptosporidium spp. and identifies gaps in our current understanding.
Keywords: Apicomplexa; Epigenetics; Non-coding RNA; Polycistronic transcription; Transcription factor.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Abrahamsen M.S., Templeton T.J., Enomoto S., Abrahante J.E., Zhu G., Lancto C.A., et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

