Genetic and clinical insights into MAST4-related neurodevelopmental disorders
- PMID: 40656202
- PMCID: PMC12245766
- DOI: 10.3389/fped.2025.1603050
Genetic and clinical insights into MAST4-related neurodevelopmental disorders
Abstract
Objective: De novo variants in MAST4 are increasingly implicated in neurodevelopmental disorders (NDDs), but the associated phenotypic spectrum remains incompletely characterized. We report a Chinese child with global developmental delay (GDD) and a novel MAST4 variant, further delineating the genotype-phenotype correlations for this gene.
Methods: Clinical and genetic data were retrospectively analyzed for a proband diagnosed with a MAST4-related NDD at Fujian Children's Hospital. Trio-based whole-exome sequencing (WES) and subsequent Sanger sequencing were performed to identify and validate the pathogenic variant.
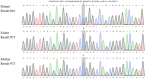
Results: The 4-year-old male proband exhibited GDD with intellectual, motor, and speech impairments. Brain MRI showed delayed myelination. WES revealed a heterozygous MAST4 missense variant (NM_001164664.2: c.4142G>T, p.Arg1381Leu), absent in population databases (gnomAD) and confirmed as de novo. The variant affects a highly conserved residue, supporting its likely pathogenicity. Phenotypic comparison with five previously reported cases confirmed core features of GDD and white matter abnormalities, though our patient lacked infantile spasms, underscoring clinical heterogeneity.
Conclusion: This study reinforces MAST4's role in NDDs and expands the genetic and phenotypic spectrum associated with this gene. The absence of infantile spasms in our case suggests variable expressivity, necessitating further functional studies to assess the variant's pathogenicity and MAST4's neurobiological mechanisms.
Keywords: MAST4; de novo variant; developmental delay; myelination dysplasia; whole-exome sequencing.
© 2025 Zheng, Fan, Lei, Xu, Peng and Zhou.
Conflict of interest statement
Author MP was employed by company Chigene (Beijing) Translational Medical Research Center Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- GBD. 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the global burden of disease study 2019. Lancet Psychiatry. (2022) 9:137–50. 10.1016/S2215-0366(21)00395-3 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

