Deciphering lactate metabolism in colorectal cancer: Prognostic modeling, immune infiltration, and gene mutation insights
- PMID: 40656609
- PMCID: PMC12243863
- DOI: 10.3748/wjg.v31.i25.107478
Deciphering lactate metabolism in colorectal cancer: Prognostic modeling, immune infiltration, and gene mutation insights
Abstract
Background: Colorectal cancer (CRC) remains a major global health burden due to its high incidence and mortality, with treatment efficacy often hindered by tumor heterogeneity, drug resistance, and a complex tumor microenvironment (TME). Lactate metabolism plays a pivotal role in reshaping the TME, promoting immune evasion and epithelial-mesenchymal transition, making it a promising target for novel therapeutic strategies and prognostic modeling in CRC.
Aim: To offer an in-depth analysis of the role of lactate metabolism in CRC, highlighting its significance in the TME and therapeutic response.
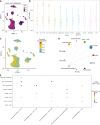
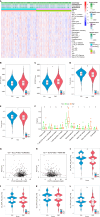
Methods: Utilizing single-cell and transcriptomic data from the Gene Expression Omnibus and The Cancer Genome Atlas, we identified key lactate metabolic activities, particularly in the monocyte/macrophage subpopulation.
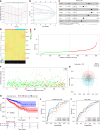
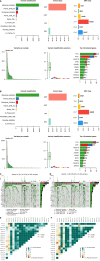
Results: Seven lactate metabolism-associated genes were significantly linked to CRC prognosis and used to construct a predictive model. This model accurately forecasts patient outcomes and reveals notable distinct patterns of immune infiltration and transcriptomic profiles mutation profiles between high- and low-risk groups. High-risk patients demonstrated elevated immune cell infiltration, increased mutation frequencies, and heightened sensitivity to specific drugs (AZD6482, tozasertib, and SB216763), providing a foundation for personalized treatment approaches. Additionally, a nomogram integrating clinical and metabolic data effectively predicted 1-, 3-, and 5-year survival rates.
Conclusion: This report underscored the pivotal mechanism of lactate metabolism in CRC prognosis and suggest novel avenues for therapeutic intervention.
Keywords: Colorectal cancer; Gene mutation analysis; Immune infiltration; Lactate metabolism; Prognostic model.
©The Author(s) 2025. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: All the authors report no relevant conflicts of interest for this article.
Figures



References
-
- Ouchi A, Shida D, Hamaguchi T, Takashima A, Ito Y, Ueno H, Ishiguro M, Takii Y, Ikeda S, Ohue M, Fujita S, Shiozawa M, Kataoka K, Ito M, Tsukada Y, Akagi T, Inomata M, Shimada Y, Kanemitsu Y. Challenges of improving treatment outcomes for colorectal and anal cancers in Japan: the Colorectal Cancer Study Group (CCSG) of the Japan Clinical Oncology Group (JCOG) Jpn J Clin Oncol. 2020;50:368–378. - PubMed
-
- Dohrn N, Klein MF. Colorectal cancer: current management and future perspectives. Br J Surg. 2023;110:1256–1259. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

