Multi-Omics Analysis Reveals the transforming growth factor-β Signaling-Driven Multicellular Interactions with Prognostic Relevance in Cervical Cancer Progression
- PMID: 40657372
- PMCID: PMC12244011
- DOI: 10.7150/jca.114505
Multi-Omics Analysis Reveals the transforming growth factor-β Signaling-Driven Multicellular Interactions with Prognostic Relevance in Cervical Cancer Progression
Abstract
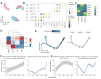
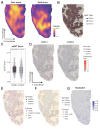
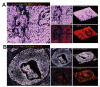
While cervical cancer (CC) prognosis depends on tumor staging, the spatiotemporal evolution of tumor microenvironment (TME) heterogeneity during metastatic progression remains poorly characterized at single-cell resolution. We employed an integrative multi-omics approach, combining single-cell RNA sequencing (scRNA-seq; n = 11), spatial transcriptomics (ST), and bulk RNA-seq data from the TCGA-CESC cohort (n = 304), to systematically map TME remodeling across CC progression stages. scRNA-seq was performed on primary lesions from patients with localized (n = 3), regional (n = 4), and metastatic (n = 4) diseases, with in-depth analyses focusing on cellular characteristics, cell type composition alterations, functional changes, differentiation trajectories, and cell-cell interaction networks. These findings were further validated using spatial transcriptomics, bulk RNA-seq data, and multiple immunohistochemistry (mIHC) experiments. ScRNA-seq data revealed that the TME of the metastatic group displayed a distinct immunosuppressive phenotype. Three key subclusters closely linked to TME remodeling in this group were identified. Notably, a novel metastasis-associated epithelial subpopulation (Epi0_AGR2), characterized by both epithelial-mesenchymal transition (EMT) and chemokine secretory phenotypes, was discovered. Gene Set Variation Analysis (GSVA) revealed that transforming growth factor β (TGF-β) signaling activation served as its primary transcriptional driver. Additionally, a neutrophil subset with pro-tumor and immunosuppressive properties, as well as a cancer-associated fibroblasts (CAFs) subset that promoted angiogenesis, were enriched in the metastatic group. Cell-cell interaction analysis and spatial mapping further revealed the formation of coordinated Epi0-neutrophil-CAFs niches, which established TGF-β-CXCL1/2/8-OSM/OSMR feedforward loops. Importantly, a computational model derived from the TME metastatic niche signature demonstrated significant prognostic stratification in the TCGA cohort (HR = 2.5179, p = 0.0144). In all, this study provides the first comprehensive delineation of stage-specific TME dynamics in CC, revealing TGF-β-driven cellular cooperativity as a metastatic switch. The joint framework establishes a potential clinically translatable tool for precision prognosis and therapeutic targeting.
Keywords: TME; cervical cancer; multi-omics analysis; niche; tumor progression.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Oaknin A, Gladieff L, Martinez-Garcia J, Villacampa G, Takekuma M, De Giorgi U. et al. Atezolizumab plus bevacizumab and chemotherapy for metastatic, persistent, or recurrent cervical cancer (BEATcc): a randomised, open-label, phase 3 trial. Lancet. 2024;403:31–43. - PubMed
-
- Wu X, Sun Y, Yang H, Wang J, Lou H, Li D. et al. Cadonilimab plus platinum-based chemotherapy with or without bevacizumab as first-line treatment for persistent, recurrent, or metastatic cervical cancer (COMPASSION-16): a randomised, double-blind, placebo-controlled phase 3 trial in China. Lancet. 2024;404:1668–76. - PubMed
-
- Lorusso D, Xiang Y, Hasegawa K, Scambia G, Leiva M, Ramos-Elias P. et al. Pembrolizumab or placebo with chemoradiotherapy followed by pembrolizumab or placebo for newly diagnosed, high-risk, locally advanced cervical cancer (ENGOT-cx11/GOG-3047/KEYNOTE-A18): a randomised, double-blind, phase 3 clinical trial. Lancet. 2024;403:1341–50. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

