Integrative proteomics and metabolomics analyses reveals the regulation of autophagy and ferroptosis by RAB10 through Slc37a2/mTOR pathway in breast cancer
- PMID: 40658144
- PMCID: PMC12259503
- DOI: 10.1007/s00018-025-05788-5
Integrative proteomics and metabolomics analyses reveals the regulation of autophagy and ferroptosis by RAB10 through Slc37a2/mTOR pathway in breast cancer
Abstract
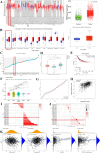
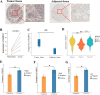
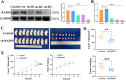
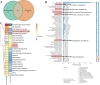
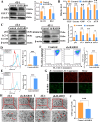
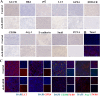
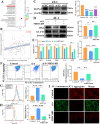
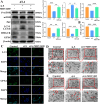
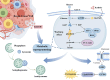
Breast cancer (BC) is the most prevalent and highly heterogeneous malignancy affecting females worldwide, and its development is closely linked to metabolic reprogramming. In this study, label-free quantification (LFQ) was used to analyze the protein expression in exosomes secreted by BC drug-resistant cells, identifying RAS-associated binding protein (RAB) 10 as the most significantly upregulated protein. RAB10, a member of the small GTPase family with complex biological functions, is highly expressed in BC and is associated with poor prognosis. In this study, we mainly utilized mouse breast cancer 4T-1 cells (wild-type control cells) and tumor-induced 4T-1 cells (isolated from mouse in situ tumor tissues to simulate the phenotype of the in vivo tumor microenvironment), and on this basis, conducted in vitro functional verification and in vivo tumorigenesis experiments. A comprehensive multi-omics analysis, including metabolomics and proteomics, following RAB10 knockdown, demonstrated the crucial role of RAB10 in regulating central carbon metabolism, which is essential for autophagy and ferroptosis in BC cells. Our study further confirmed that RAB10 mediates metabolic reprogramming in BC cells by regulating the Slc37a2/mTOR pathway, leading to enhanced autophagy and inhibition of ferroptosis. This comprehensive multi-omics analysis elucidated the key molecular and regulatory mechanisms underlying RAB10-induced metabolic reprogramming in tumors, providing potential new therapeutic targets and biomarkers for prognostic assessment in BC treatment.
Keywords: Autophagy; Breast cancer; Ferroptosis; Metabolism; RAB10.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics statement: All animal experiments were performed under the policy of the Ethics Committee of Bengbu Medical University. The study was approved by the Ethics Committee of Bengbu Medical University and the Ethics Committee of Laboratory Animal Management (Ethics No.: LENKO Approval No. [2023] 537). Conflict of interest: The authors declare no conflict of interest.
Figures


Similar articles
-
Upregulated SAE1 Drives Tumorigenesis and Is Associated with Poor Clinical Outcomes in Breast Cancer.Breast J. 2024 Jun 30;2024:2981722. doi: 10.1155/2024/2981722. eCollection 2024. Breast J. 2024. PMID: 39742381 Free PMC article.
-
M1 Macrophage-Derived TNF-α Promotes Pancreatic Cancer Ferroptosis Via p38 MAPK-ACSL4 Pathway.Curr Mol Med. 2025 Jul 10. doi: 10.2174/0115665240374551250630075409. Online ahead of print. Curr Mol Med. 2025. PMID: 40653839
-
Glucose-6-phosphate dehydrogenase (G6PD) shields pancreatic cancer from autophagy-dependent ferroptosis by suppressing redox imbalance induced AMPK/mTOR signaling.Free Radic Biol Med. 2025 Sep;237:195-209. doi: 10.1016/j.freeradbiomed.2025.06.002. Epub 2025 Jun 2. Free Radic Biol Med. 2025. PMID: 40466783
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Bray F, Laversanne M et al (2024) Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J/OL]. CA: a cancer journal for clinicians. 74(3):229–263. 10.3322/caac.21834 - PubMed
-
- Chua CEL, Tang BL (2018) Rab 10-a traffic controller in multiple cellular pathways and locations[J/OL]. J Cell Physiol 233(9):6483–6494. 10.1002/jcp.26503 - PubMed
-
- Liu J, Bai X, Zhang M et al (2024) Energy metabolism: a new target for gastric cancer treatment[J/OL]. Clinical & translational oncology: official publication of the federation of Spanish oncology societies and of the National Cancer Institute of Mexico. 26(2):338–351. 10.1007/s12094-023-03278-3 - PubMed
MeSH terms
Substances
Grants and funding
- GXXT-2022-064/the University Synergy Innovation Program of Anhui Province
- 2024AH010021/excellent Scientific Research and Innovation Team of Anhui Universities
- No.: KJ2019ZD28/the Major Program of Anhui Educational Committee
- Byycx22042, Byycx23013,Byycx24035/the Program for graduate's research of Bengbu Medical College
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

