Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes
- PMID: 40659618
- PMCID: PMC12259956
- DOI: 10.1038/s41467-025-60251-0
Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes
Abstract
Regeneration and homeostatic turnover of solid tissues depend on the proliferation of symmetrically dividing adult stem cells, which either remain stem cells or differentiate based on their niche position. Here we demonstrate that in zebrafish lateral line sensory organs, stem and progenitor cell proliferation are independently regulated by two cyclinD genes. Loss of ccnd2a impairs stem cell proliferation during development, while loss of ccndx disrupts hair cell progenitor proliferation but allows normal differentiation. Notably, ccnd2a can functionally replace ccndx, indicating that the respective effects of these Cyclins on proliferation are due to cell type-specific expression. However, even though hair cell progenitors differentiate normally in ccndx mutants, they are mispolarized due to hes2 and Emx2 downregulation. Thus, regulated proliferation ensures that equal numbers of hair cells are polarized in opposite directions. Our study reveals cell type-specific roles for cyclinD genes in regulating the different populations of symmetrically dividing cells governing organ development and regeneration, with implications for regenerative medicine and disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
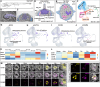


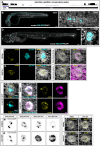



Update of
-
Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes.bioRxiv [Preprint]. 2024 Oct 22:2024.10.21.619490. doi: 10.1101/2024.10.21.619490. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 14;16(1):5913. doi: 10.1038/s41467-025-60251-0. PMID: 39484411 Free PMC article. Updated. Preprint.
Similar articles
-
Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes.bioRxiv [Preprint]. 2024 Oct 22:2024.10.21.619490. doi: 10.1101/2024.10.21.619490. bioRxiv. 2024. Update in: Nat Commun. 2025 Jul 14;16(1):5913. doi: 10.1038/s41467-025-60251-0. PMID: 39484411 Free PMC article. Updated. Preprint.
-
Semaphorin 3f and post-embryonic regulation of retinal progenitors.PLoS Genet. 2025 Jul 14;21(7):e1011748. doi: 10.1371/journal.pgen.1011748. eCollection 2025 Jul. PLoS Genet. 2025. PMID: 40658724 Free PMC article.
-
Zebrafish Kelch-like family member 4 is required for vasculogenesis and hematopoiesis.Dev Biol. 2025 Sep;525:1-12. doi: 10.1016/j.ydbio.2025.05.018. Epub 2025 May 20. Dev Biol. 2025. PMID: 40404079
-
Relationship between Pituitary Gland and Stem Cell in the Aspect of Hormone Production and Disease Prevention: A Narrative Review.Endocr Metab Immune Disord Drug Targets. 2025;25(7):509-526. doi: 10.2174/0118715303314551241031093717. Endocr Metab Immune Disord Drug Targets. 2025. PMID: 39812047 Review.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
References
-
- Doupe, D. P., Klein, A. M., Simons, B. D. & Jones, P. H. The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell18, 317–323 (2010). - PubMed
-
- Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell143, 134–144 (2010). - PubMed
-
- Lopez-Garcia, C., Klein, A. M., Simons, B. D. & Winton, D. J. Intestinal stem cell replacement follows a pattern of neutral drift. Science330, 822–825 (2010). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

