Quantitative phase imaging with temporal kinetics predicts hematopoietic stem cell diversity
- PMID: 40659629
- PMCID: PMC12260078
- DOI: 10.1038/s41467-025-61846-3
Quantitative phase imaging with temporal kinetics predicts hematopoietic stem cell diversity
Abstract
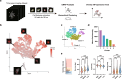
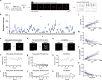
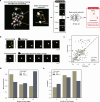
Innovative identification technologies for hematopoietic stem cells (HSCs) have expanded the scope of stem cell biology. Clinically, the functional quality of HSCs critically influences the safety and therapeutic efficacy of stem cell therapies. However, most analytical techniques capture only a single snapshot, disregarding the temporal context. A comprehensive understanding of the temporal heterogeneity of HSCs necessitates live-cell, real-time and non-invasive analysis. Here, we developed a prediction system for HSC diversity by integrating single-HSC ex vivo expansion technology with quantitative phase imaging (QPI)-driven machine learning. By analyzing the cellular kinetics of individual HSCs, we discovered previously undetectable diversity that snapshot analysis cannot resolve. The QPI-driven algorithm quantitatively evaluates stemness at the single-cell level and leverages temporal information to significantly improve prediction accuracy. This platform advances the field from snapshot-based identification of HSCs to dynamic, time-resolved prediction of their functional quality based on past cellular kinetics.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- TILL, J. E. & McCULLOCH, E. A. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res.14, 213–222 (1961). - PubMed
-
- Spangrude, G. J., Heimfeld, S. & Weissman, I. L. Purification and characterization of mouse hematopoietic stem cells. Science241, 58–62 (1988). - PubMed
-
- Morrison, S. J. & Weissman, I. L. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by phenotype. Immunity1, 661–673 (1994). - PubMed
-
- Osawa, M., Hanada, K. I., Hamada, H. & Nakauchi, H. Long-term lymphohematopoietic reconstitution by a single CD34- low/negative hematopoietic stem cell. Science273, 242–245 (1996). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

