Boosting RNA nanotherapeutics with V-ATPase activating non-inflammatory lipid nanoparticles to treat chronic lung injury
- PMID: 40659631
- PMCID: PMC12259844
- DOI: 10.1038/s41467-025-61688-z
Boosting RNA nanotherapeutics with V-ATPase activating non-inflammatory lipid nanoparticles to treat chronic lung injury
Abstract
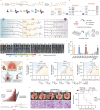
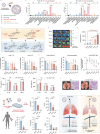
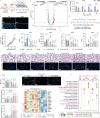
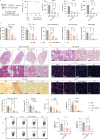
Lipid nanoparticles (LNPs) are a promising platform for mRNA delivery. However, their use in inflammatory pulmonary diseases is limited by reactogenicity and suboptimal delivery. Here we develop a non-inflammatory LNP (NIF-LNP) by incorporating ursolic acid, identified from a natural product library, into a biodegradable, cationic phosphoramide-derived LNP formulation. NIF-LNPs exhibit a 40-fold enhancement in lung protein expression without causing significant reactogenicity compared to LNPs containing ALC-0315. Our CRISPR-KO mechanistic studies uncover that ursolic acid promote endosome acidification by activating the V-ATPase complex, acting as a central hub for endosomal trafficking of LNPs and inflammation control. Furthermore, we identify an intracellular circadian regulatory gene, NR1D1, encapsulated in NIF-LNPs, showing notable therapeutic efficacy in bronchopulmonary dysplasia and lung fibrosis. To enhance clinical feasibility, we have developed a lyophilized formulation that maintains stability for over 90 days and ensures efficient nebulization in preclinical male mouse, pup rat, and male dog models. Overall, this V-ATPase-activating atomized NIF-LNP presents a viable strategy for treating variable chronic inflammatory lung diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: L.M., Z.Q.Z. and X.Z.S. have filed a patent for the development of the described NIF LNPs. The remaining authors declare no competing interests.
Figures


References
-
- Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol.37, 1174 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- Z220022/Natural Science Foundation of Beijing Municipality (Beijing Natural Science Foundation)
- Z231100007223012/Beijing Municipal Science and Technology Commission
- 82373807, 82070460, 82270479, 32030064, 32250013, 22175188/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

