Immunodiagnostic plasma amino acid residue biomarkers detect cancer early and predict treatment response
- PMID: 40659634
- PMCID: PMC12260101
- DOI: 10.1038/s41467-025-61685-2
Immunodiagnostic plasma amino acid residue biomarkers detect cancer early and predict treatment response
Abstract
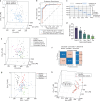
The immune response to tumour development is frequently targeted with therapeutics but remains largely unexplored in diagnostics, despite being stronger for early-stage tumours. We present an immunodiagnostic platform to detect this. We identify a panel of amino acid residue biomarkers providing a signature of cancer-specific immune activation associated with tumour development and distinct from autoimmune and infectious diseases, measurable optically in neat blood plasma, and validate within N = 170 participants. By measuring the total concentrations of cysteine, free cysteine, lysine, tryptophan, and tyrosine protein-incorporated biomarkers and analyzing the results with supervised machine learning, we identify 78% of cancers with 0% false positive rate (N = 97) with an AUROC of 0.95. The cancer, healthy, and autoimmune/infectious biomarker pattern are statistically significantly different (p < 0.0001). Smaller-scale changes in biomarker concentrations reveal inter-patient differences in immune activation that predict treatment response. Specific concentration ranges of these biomarkers predict response to Cyclin-dependent kinase inhibitors in advanced breast cancer patients (p < 0.05), identifying 98% of responders (N = 33). Here we provide an immunodiagnostic technology platform that, to our knowledge, has not been previously reported, and prove initial clinical application in a cohort of N = 170, including proof of concept in Multi Cancer Early Detection and personalized medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: W.S., E.V.Y. and G.J.L.B. are co-founders of Proteotype Diagnostics Ltd. C.T., W.S., L.C., E.V.Y. and G.J.L.B. are stockholders of Proteotype Diagnostics Ltd. W.S. and E.V.Y. are employed by Proteotype Diagnostics Ltd. Proteotype Diagnostics Ltd owns a patent application that incorporates a method of identifying the presence and/or concentration and/or amount of proteins or proteomes, which is described in this manuscript (EP4196797A1). G.J.L.B. is a Visiting Professor at Xi’an Fengcheng Hospital. All other authors declare no conflict of interest.
Figures




References
-
- Dagher, O. K., Schwab, R. D., Brookens, S. K. & Posey, A. D. Advances in cancer immunotherapies. Cell186, 1814–1814.e1 (2023). - PubMed
-
- Nicholson, B. D. et al. Multi-cancer early detection test in symptomatic patients referred for cancer investigation in England and Wales (SYMPLIFY): a large-scale, observational cohort study. Lancet Oncol.24, 733–743 (2023). - PubMed
-
- Klein, E. et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol.32, 1167–1177 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

