Atlas of amnion development during the first trimester of human pregnancy
- PMID: 40659869
- PMCID: PMC12270915
- DOI: 10.1038/s41556-025-01696-9
Atlas of amnion development during the first trimester of human pregnancy
Abstract
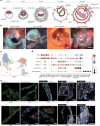
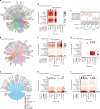
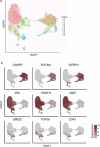
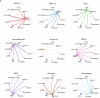
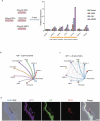
The amnion is a critical extra-embryonic structure that supports foetal development, yet its ontogeny remains poorly defined. Here, using single-cell transcriptomics, we identified major cell types and subtypes in the human amnion across the first trimester of pregnancy, broadly categorized into epithelial, mesenchymal and macrophage lineages. We uncovered epithelial-mesenchymal and epithelial-immune transitions, highlighting dynamic remodelling during early pregnancy. Our results further revealed key intercellular communication pathways, including BMP4 signalling from mesenchymal to epithelial cells and TGF-β signalling from macrophages to mesenchymal cells, suggesting coordinated interactions that drive amnion morphogenesis. In addition, integrative comparisons across humans, non-human primates and in vitro stem cell-based models reveal that stem cell-based models recapitulate various stages of amnion development, emphasizing the need for careful selection of model systems to accurately recapitulate in vivo amnion formation. Collectively, our findings provide a detailed view of amnion cellular composition and interactions, advancing our understanding of its developmental role and regenerative potential.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








References
-
- Mole, M. A., Weberling, A. & Zernicka-Goetz, M. Comparative analysis of human and mouse development: from zygote to pre-gastrulation. Curr. Top. Dev. Biol.136, 113–138 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

