Molecular characterization of ten viral pathogens causing calf diarrhea in Hanwoo (Bos Taurus coreanae) by next generation sequencing
- PMID: 40660160
- PMCID: PMC12257796
- DOI: 10.1186/s12917-025-04926-2
Molecular characterization of ten viral pathogens causing calf diarrhea in Hanwoo (Bos Taurus coreanae) by next generation sequencing
Abstract
Background: Calf diarrhea remains a significant concern in the global cattle industry, leading to considerable economic losses. Infectious pathogens are among the primary causes of this disease. In this study, the prevalence of 7 pathogens—bovine rotavirus (BRV), bovine coronavirus (BCV), bovine viral diarrhea virus (BVDV) types 1 and 2, Cryptosporidium parvum, Giardia spp., and Eimeria spp.—associated with calf diarrhea was investigated using polymerase chain reaction (PCR). A metagenomic approach was also applied to identify additional RNA viral pathogens from unknown causes of diarrheic fecal samples in the Republic of Korea (ROK).
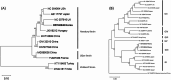
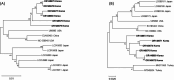
Results: A total of 810 fecal samples from Hanwoo calves (Bos taurus coreanae) were collected, consisting of 526 normal samples (267 with a fecal score of 0 and 259 with a fecal score of 1) and 284 diarrheic samples (178 with a fecal score of 2 and 106 with a fecal score of 3). All 7 pathogens were detected by PCR in feces and their detection rates and mean fecal scores for each were as follows: BRV (14.0%, 1.41), BCV (3.2%, 1.42), BVDV1 (2.1%, 1.35), BVDV2 (4.9%, 1.33), C. parvum (9.8%, 1.66), Eimeria spp. (1.9%, 1.73), and Giardia spp. (0.9%, 0.71). Among these pathogens, BRV (p = 0.004), C. parvum (p < 0.001), and Eimeria spp. (p = 0.027) showed an increase in prevalence with higher fecal scores. Twenty-one fecal samples negative for all pathogens were randomly selected and subjected to high-throughput sequencing to identify RNA viral pathogens associated with calf diarrhea. This approach led to the identification of nearly complete genomic sequences for bovine astrovirus, bovine enterovirus, bovine kobuvirus, bovine nebovirus, bovine norovirus, bovine boosepivirus B, bovine parechovirus, bovine torovirus, C. parvum virus 1, and hunnivirus.
Conclusions: This study represents the first investigation of hunnivirus presence and provides a comprehensive description of the nearly complete genomes of 10 viruses associated with calf diarrhea in the ROK. The findings contribute to a better understanding of the epidemiology and molecular characteristics of calf diarrhea-associated pathogens in the ROK, highlighting the potential application of high-throughput sequencing for diagnosing other diseases.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12917-025-04926-2.
Keywords: Calf diarrhea; Hanwoo calves; Next-generation sequencing; Phylogenetic analysis; Prevalence; Viral detection.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical review and approval were waived for this study because it does not involve any studies with live animals conducted by any of the authors. All samples used in this study were received from farmers or field veterinarians who submitted the samples for diagnostic purposes. Consent for publication: An informed consent statement was provided by all animal owners. Competing interests: The authors declare no competing interests.
Figures



References
-
- Singh BB, Sharma R, Kumar H, Banga HS, Aulakh RS, Gill JPS, Sharma JK. Prevalence of Cryptosporidium parvum infection in Punjab (India) and its association with diarrhea in neonatal dairy calves. Vet Parasitol. 2006;140(1):162–5. - PubMed
Grants and funding
- 122061-2/Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry
- 122061-2/Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry
- 122061-2/Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry
LinkOut - more resources
Full Text Sources
Miscellaneous

