Chemoresistant tumor cell secretome potentiates immune suppression in triple negative breast cancer
- PMID: 40660361
- PMCID: PMC12257726
- DOI: 10.1186/s13058-025-02082-x
Chemoresistant tumor cell secretome potentiates immune suppression in triple negative breast cancer
Abstract
Background: Chemotherapy is an integral part of the clinical management of triple negative breast cancer (TNBC), however, development of chemoresistance occurs frequently. Tumor sensitivity to treatment is known to be strongly influenced by the immune microenvironment, signifying the predictive potential of immune alterations. How tumor cells that acquire resistance may subsequently modulate the immune microenvironment it is still not well described. Here, we investigated immunomodulation in the context of acquired chemoresistance in TNBC, focusing on the role of the secretome.
Methods: Bulk RNA sequencing and multiplex cytokine profiling were performed on paclitaxel-resistant and -sensitive isogenic variants of TNBC cells to reveal resistance-associated secretome alterations. The immunomodulatory influence of the tumor cell secretome was investigated by exploring its effect on monocytes, macrophages (MΦs) and T cells derived from healthy blood donors. The influence on the immune cell phenotype and activity was evaluated by measuring molecular markers and performing functional assays. To validate the clinical relevance, we utilized longitudinal -omics data from breast cancer patients refractory to standard chemotherapy in the NeoAva clinical trial. CIBERSORT was applied to transcriptomics data to infer MΦ and T cell abundance in individual tumors upon treatment. To evaluate their association with the secretome profiles, patient-matched serum cytokine data were used.
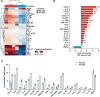
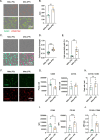
Results: The acquisition of chemoresistance was accompanied by enhanced secretion of cytokines. Subsequently, the resistant cell secretome affected the abundance, phenotype and activity of immune cells. Specifically, it potentiated the recruitment of monocytes, facilitated the polarization of MΦs towards the immunosuppressive M2-like phenotype, and attenuated the activation of CD8+ T cells. Data from the NeoAva clinical cohort validated the enrichment of M2 MΦs and/or the depletion of M1 MΦs after treatment in the majority of residual tumors. The MΦ-associated changes counteracted CD8+ T cell abundance and were partially associated with the cytokine-enriched secretome.
Conclusion: Development of chemoresistance in BC is associated with alterations in the tumor secretome, which impairs immune activation and facilitates immunosuppression. Knowledge on the immune microenvironment in residual tumors after standard chemotherapy could aid in selecting rational treatment options for this group of patients.
Keywords: Chemoresistance; Cytokines; Immune microenvironment; Immune suppression; Macrophages; Secretome; Triple negative breast cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approvals were obtained from the Norwegian Regional Committee for Medical and Health Research Ethics (REK) for experiments using monocytes isolated from buffy coats of healthy anonymous donors (REK 95986) and for T cell isolation and generation of TCR-engineered T cells (REK 2019–121). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
-
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15):4429. - PubMed
-
- Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164. - PubMed
-
- Ladoire S, Mignot G, Dabakuyo S, Arnould L, Apetoh L, Rébé C, et al. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. J Pathol. 2011;224(3):389. - PubMed
MeSH terms
Substances
Grants and funding
- 859962/European Union´s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement
- 2021049/South-East Norway Regional Health Authorities
- 2022069/South-East Norway Regional Health Authorities
- 847912/European Union Horizon 2020 research and innovation program
- 190257-2017/Norwegian Cancer Society
LinkOut - more resources
Full Text Sources
Research Materials

