This is a preprint.
Autocrine TGFβ2 enforces a transcriptionally hybrid cell state in Ewing sarcoma
- PMID: 40661358
- PMCID: PMC12259098
- DOI: 10.1101/2025.06.11.653876
Autocrine TGFβ2 enforces a transcriptionally hybrid cell state in Ewing sarcoma
Abstract
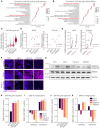
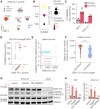
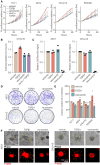
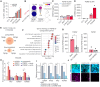
Sub-populations of cancer-associated fibroblast (CAF)-like tumor cells deposit extracellular matrix (ECM) proteins that support Ewing sarcoma (EwS) progression and metastasis. We previously showed a hallmark of CAF-like EwS cells is their hybrid transcriptional state wherein the driver fusion oncogene, EWS::FLI1, maintains activation of proliferative programs but loses capacity to repress mesenchymal genes. Here, we studied primary patient tumors and cell line models to identify molecular drivers of this hybrid state. Our data reveal that hybrid EwS cells are induced and maintained by a TGFβ signaling positive feedback loop. Hybrid cells de-repress TGFBR2 and upregulate expression and secretion of TGFβ2 to sustain pathway activation and ECM deposition. While TGFβ ligands can potently induce growth arrest in cells of epithelial origin, we show that TGFβ1 and TGFβ2 promote cell invasion of EwS cells without affecting proliferation. Thus, stroma and tumor-derived TGFβ ligands induce and maintain hybrid EwS cells to promote pro-metastatic cell phenotypes.
Conflict of interest statement
Competing Interests: KMB: DSMC member for Merck. All other authors declare that they have no competing interests.
Figures

References
-
- Hanahan D., Hallmarks of Cancer: New Dimensions. Cancer Discovery 12, 31–46 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
