This is a preprint.
Filopodia numbers impact chemotactic migration speed
- PMID: 40661508
- PMCID: PMC12258711
- DOI: 10.1101/2025.05.28.656686
Filopodia numbers impact chemotactic migration speed
Abstract
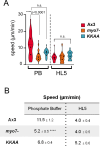
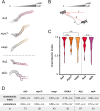
Migrating cells sense and respond to external chemical and physical cues, enabling them to efficiently reach their destinations. Filopodia are slender actin-filled membrane protrusions implicated in interacting with the extracellular environment in many contexts, such as neuronal growth cone guidance and the capture of prey by immune cells and unicellular organisms. The role of filopodia in chemotactic guidance in fast-moving amoeboid cells has not been well-studied. The social amoeba Dictyostelium relies on chemotaxis for development and finding food, making it an excellent system for investigating the role of filopodia in amoeboid chemotaxis. Stimulation of amoebae with the chemoattractant cAMP activates a transient increase of filopodia formation by recruiting the filopodial myosin DdMyo7 to the cell cortex. Filopodia formation is biased towards the source of chemoattractant, yet myo7 null cells that lack filopodia exhibit normal directional migration. However, cells either lacking filopodia or having increased numbers of filopodia move more slowly than those with wildtype numbers of filopodia. Thus, while filopodia are dispensable for detection of chemical gradients by amoeboid cells, changes in filopodia number can impact their migration speed possibly due to altering cell-substrate adhesion.
Keywords: actin; amoeboid migration; chemotaxis; directed migration; filopodia; myosin.
Figures



Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Modulation of Filopodial Myosin Function.bioRxiv [Preprint]. 2025 May 29:2025.05.29.656896. doi: 10.1101/2025.05.29.656896. bioRxiv. 2025. PMID: 40501999 Free PMC article. Preprint.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
References
-
- Bentley D, Toroian-Raymond A (1986). Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature 323, 712–715. - PubMed
-
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M et al. (2019). ilastik: interactive machine learning for (bio)image analysis. Nat Meth 16, 1226–1232. - PubMed
-
- Brazill D (2016). Chemotaxis: Under Agarose Assay. In Cytoskeleton: Methods and Protocols, Gavin RH, ed. (New York: Springer Science + Business Media; ), pp. 339–346.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
