This is a preprint.
Multi-selective RAS(ON) Inhibition Targets Oncogenic RAS Mutations and Overcomes RAS/MAPK-Mediated Resistance to FLT3 and BCL2 Inhibitors in Acute Myeloid Leukemia
- PMID: 40661530
- PMCID: PMC12259086
- DOI: 10.1101/2025.06.10.658786
Multi-selective RAS(ON) Inhibition Targets Oncogenic RAS Mutations and Overcomes RAS/MAPK-Mediated Resistance to FLT3 and BCL2 Inhibitors in Acute Myeloid Leukemia
Abstract
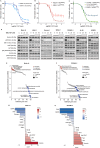
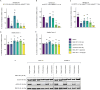
Aberrant activation of the RAS/MAPK signaling limits the clinical efficacy of several targeted therapies in acute myeloid leukemia (AML). In FLT3-mutant AML, the selection of clones harboring heterogeneous RAS mutations drives resistance to FLT3 inhibitors (FLT3i). RAS activation is also associated with resistance to other AML targeted therapies, including the BCL2 inhibitor venetoclax. Despite the critical need to inhibit RAS/MAPK signaling in AML, no targeted therapies have demonstrated clinical benefit in RAS-driven AML. To address this unmet need, we investigated the preclinical activity of RMC-7977, a multi-selective inhibitor of GTP-bound active [RAS(ON)] isoforms of mutant and wild-type RAS in AML models. RMC-7977 exhibited potent antiproliferative and pro-apoptotic activity across AML cell lines with MAPK-activating signaling mutations. In cell line models with acquired FLT3i resistance due to secondary RAS mutations, treatment with RMC-7977 restored sensitivity to FLT3i. Similarly, RMC-7977 effectively reversed resistance to venetoclax in RAS-addicted cell line models with both RAS wild-type and mutant genetic backgrounds. In murine patient-derived xenograft models of RAS-mutant AML, RMC-7977 was well tolerated and significantly suppressed leukemic burden in combination with gilteritinib or venetoclax. Our findings strongly support clinical investigation of broad-spectrum RAS(ON) inhibition in AML to treat and potentially prevent drug resistance due to activated RAS signaling.
Conflict of interest statement
C.C.S. reports research funding from Revolution Medicines, ERASCA, Abbvie; funding for clinical trials from Zentalis and Biomea; has served on advisory boards for Abbvie, Genentech, Servier and Biomea, and as a consultant for Astellas.
Figures




References
-
- Cortes JE, Khaled S, Martinelli G, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20(7):984–997. doi: 10.1016/S1470-2045(19)30150-0 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
