This is a preprint.
Single-cell Analysis of Intracellular Transport and Expression of Cell Surface Proteins
- PMID: 40661627
- PMCID: PMC12259062
- DOI: 10.1101/2025.06.09.658704
Single-cell Analysis of Intracellular Transport and Expression of Cell Surface Proteins
Abstract
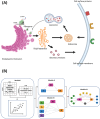
Intracellular protein transport (ICT) is a tightly regulated process that orchestrates protein localization and expression, ensuring proper cellular function. Dysregulated ICT can lead to aberrant expression of surface proteins involved in cell-cell communication, adhesion, and immune responses, contributing to disease progression and therapeutic resistance. Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) enables the simultaneous measurement of mRNA and surface protein levels in the same cell, providing a powerful opportunity to investigate the molecular mechanisms underlying surface protein regulation. In this study, we introduce a novel computational frame for Modeling Protein Expression and Transport (MPET) that evaluates the contribution of ICT activity to differential surface protein expression using CITE-seq data. MPET comprises three modules for identification of ICT-surface protein regulatory circuits across biological scales and their contributions to phenotypic variation. We applied MPET to analyze single-cell data from COVID-19 patients with varying disease severity. Our analysis revealed context-dependent recruitment of ICT genes and pervasive rewiring of ICT pathways throughout the course of disease progression. Notably, we found that even when the transcriptional levels of key immune response proteins remained stable, their expression on cell surface were significantly altered due to dysregulated ICT. MPET provides a valuable new tool for dissecting complex regulatory networks and offers mechanistic insight into post-transcriptional regulation of cell surface proteins in diseases.
Keywords: CITE-seq; COVID-19; immune dysregulation; intracellular transport; mediation analysis; mixed-effects modeling; post-transcriptional regulation; single-cell multi-omics; surface protein expression.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures





References
-
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P. (2002). Membrane Proteins.
-
- Xiong S., and Xu W. (2014). Human Immune System. Comprehensive Biomedical Physics 10, 91–114. 10.1016/B978-0-444-53632-7.01008-X. - DOI
-
- Engel P., Boumsell L., Balderas R., Bensussan A., Gattei V., Horejsi V., Jin B.-Q., Malavasi F., Mortari F., Schwartz-Albiez R., et al. (2015). CD Nomenclature 2015: Human Leukocyte Differentiation Antigen Workshops as a Driving Force in Immunology. The Journal of Immunology 195, 4555–4563. 10.4049/JIMMUNOL.1502033. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
