LncRNA NEAT1 promotes epithelial-mesenchymal transition in nasal polyp cells via the miR-199-3p/PAK4 axis
- PMID: 40661947
- PMCID: PMC12256258
- DOI: 10.3389/fimmu.2025.1613179
LncRNA NEAT1 promotes epithelial-mesenchymal transition in nasal polyp cells via the miR-199-3p/PAK4 axis
Abstract
Background and purpose: Chronic rhinosinusitis with nasal polyps (CRSwNP) is a persistent inflammatory condition marked by high recurrence and limited therapeutic efficacy. This study investigates the role of long non-coding RNA NEAT1 in promoting epithelial-mesenchymal transition (EMT) in CRSwNP, focusing on its regulatory interaction with the miR-199-3p/PAK4 axis.
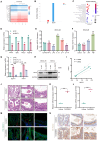
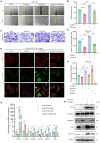
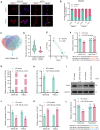
Methods: NEAT1 expression was assessed in nasal epithelial cells from CRSwNP patients using qPCR and FISH. Primary human nasal epithelial cells and BEAS-2B cells were subjected to NEAT1 knockdown via siRNA. Cell migration, barrier function, and cytoskeletal dynamics were evaluated through scratch assays, Transwell migration, FITC-Dextran permeability testing, and phalloidin staining. EMT marker expression was analyzed via Western blotting and immunofluorescence. Transcriptome sequencing identified PAK4 as a downstream effector. In vivo validation was performed using a mouse nasal polyp model, and molecular interactions among NEAT1, miR-199-3p, and PAK4 were confirmed via dual-luciferase reporter assays. Rescue experiments further elucidated mechanistic pathways.
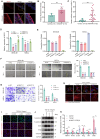
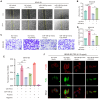
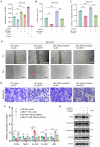
Results: In comparison to controls, NEAT1 expression was significantly elevated in the epithelial tissues of CRSwNP. NEAT1 knockdown inhibited cell migration, enhanced epithelial barrier integrity, and reversed EMT-associated cytoskeletal remodeling. E-cadherin levels increased, while N-cadherin and vimentin decreased. Transcriptomic and functional analyses identified PAK4 as a NEAT1-regulated target. NEAT1 was shown to sponge miR-199-3p, thereby relieving its inhibitory effect on PAK4. Overexpression of miR-199-3p suppressed PAK4 and mitigated EMT-related changes induced by NEAT1.
Conclusion: NEAT1 promotes EMT in nasal polyp epithelial cells by modulating the miR-199-3p/PAK4 axis, highlighting its potential as a diagnostic biomarker and therapeutic target in CRSwNP.
Keywords: CRSwNP; EMT; NEAT1; PAK4; miR-199-3p; non-coding RNA.
Copyright © 2025 Li, Jiang, Zhang, Zheng, Yuan, Shen, Zhao, Lu and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Osthole Ameliorates Cigarette Smoke-Induced Epithelial-To-Mesenchymal Transition via PI3K/Akt/NF-κB Pathway in Chronic Rhinosinusitis with Nasal Polyps.Int Arch Allergy Immunol. 2025;186(8):747-757. doi: 10.1159/000543408. Epub 2025 Jan 3. Int Arch Allergy Immunol. 2025. PMID: 39756395
-
Expression of lncRNA NEAT1 in endometriosis and its biological functions in ectopic endometrial cells as mediated via miR-124-3p.Genes Genomics. 2022 May;44(5):527-537. doi: 10.1007/s13258-021-01184-y. Epub 2022 Jan 30. Genes Genomics. 2022. PMID: 35094286
-
MiR-221-3p Attenuates IL-33-Induced Mast Cell Cytokine Expression by Targeting KIT.Int Forum Allergy Rhinol. 2025 Aug;15(8):837-850. doi: 10.1002/alr.23558. Epub 2025 Mar 25. Int Forum Allergy Rhinol. 2025. PMID: 40131162 Free PMC article.
-
Periostin as a biomarker in chronic rhinosinusitis: A contemporary systematic review.Int Forum Allergy Rhinol. 2022 Dec;12(12):1535-1550. doi: 10.1002/alr.23018. Epub 2022 May 30. Int Forum Allergy Rhinol. 2022. PMID: 35514144
-
Systemic and topical antibiotics for chronic rhinosinusitis.Cochrane Database Syst Rev. 2016 Apr 26;4(4):CD011994. doi: 10.1002/14651858.CD011994.pub2. Cochrane Database Syst Rev. 2016. PMID: 27113482 Free PMC article.
References
-
- Papacharalampous GX, Constantinidis J, Fotiadis G, Zhang N, Bachert C, Katotomichelakis M. Chronic rhinosinusitis with nasal polyps (CRSwNP) treated with omalizumab, dupilumab, or mepolizumab: A systematic review of the current knowledge towards an attempt to compare agents’ efficacy. Int Forum Allergy Rhinol. (2024) 14:96–109. doi: 10.1002/alr.23234 - DOI - PubMed
-
- Bachert C, Han JK, Wagenmann M, Hosemann W, Lee SE, Backer V, et al. EUFOREA expert board meeting on uncontrolled severe chronic rhinosinusitis with nasal polyps (CRSwNP) and biologics: Definitions and management. J Allergy Clin Immunol. (2021) 147:29–36. doi: 10.1016/j.jaci.2020.11.013 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

