HIF-1 promotes murine breast cancer brain metastasis by increasing production of integrin β3-containing extracellular vesicles
- PMID: 40662366
- PMCID: PMC12259260
- DOI: 10.1172/JCI190470
HIF-1 promotes murine breast cancer brain metastasis by increasing production of integrin β3-containing extracellular vesicles
Abstract
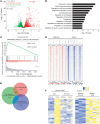
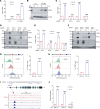
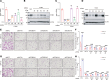
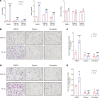
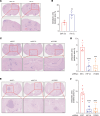
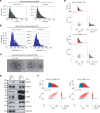
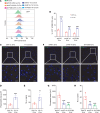
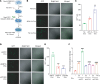
Brain metastasis is a major cause of breast cancer (BC) mortality, but the cellular and molecular mechanisms have not been fully elucidated. BC cells must breach the blood-brain barrier in order to colonize the brain. Here, we determined that integrin β3 (ITGB3) expression mediated by hypoxia-inducible factor 1 (HIF-1) plays a critical role in metastasis of BC cells to the brain. Hypoxia stimulated BC cell migration and invasion ex vivo and brain colonization in vivo. Knockdown of either HIF-1α or ITGB3 expression impaired brain colonization by human or mouse BC cells injected into the cardiac left ventricle. Exposure of BC cells to hypoxia increased expression of ITGB3 and its incorporation into small extracellular vesicles (EVs). EVs harvested from the conditioned medium of hypoxic BC cells showed increased retention in the brain after intracardiac injection that was HIF-1α and ITGB3 dependent. EVs from hypoxic BC cells showed binding to brain endothelial cells (ECs), leading to increased EC-BC cell interaction, increased vascular endothelial growth factor receptor 2 signaling, increased EC permeability, and increased transendothelial migration of BC cells. Taken together, our studies implicate HIF-1-stimulated production of ITGB3+ EVs as a key mechanism by which hypoxia promotes BC brain metastasis.
Keywords: Breast cancer; Hypoxia; Oncology; Vascular biology.
Conflict of interest statement
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

