ARTEMIS integrates autoencoders and Schrödinger Bridges to predict continuous dynamics of gene expression, cell population, and perturbation from time-series single-cell data
- PMID: 40662824
- PMCID: PMC12261462
- DOI: 10.1093/bioinformatics/btaf218
ARTEMIS integrates autoencoders and Schrödinger Bridges to predict continuous dynamics of gene expression, cell population, and perturbation from time-series single-cell data
Abstract
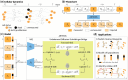
Summary: Cellular processes like development, differentiation, and disease progression are highly complex and dynamic (e.g. gene expression). These processes often undergo cell population changes driven by cell birth, proliferation, and death. Single-cell sequencing enables gene expression measurement at the cellular resolution, allowing us to decipher cellular and molecular dynamics underlying these processes. However, the high costs and destructive nature of sequencing restrict observations to snapshots of unaligned cells at discrete timepoints, limiting our understanding of these processes and complicating the reconstruction of cellular trajectories. To address this challenge, we propose ARTEMIS, a generative model integrating a variational autoencoder (VAE) with unbalanced Diffusion Schrödinger Bridge to model cellular processes by reconstructing cellular trajectories, reveal gene expression dynamics, and recover cell population changes. The VAE maps input time-series single-cell data to a continuous latent space, where trajectories are reconstructed by solving the Schrödinger bridge problem using forward-backward stochastic differential equations (SDEs). A drift function in the SDEs captures deterministic gene expression trends. An additional neural network estimates time-varying kill rates for single cells along trajectories, enabling recovery of cell population changes. Using three scRNA-seq datasets-pancreatic β-cell differentiation, zebrafish embryogenesis, and epithelial-mesenchymal transition (EMT) in cancer cells-we demonstrate that ARTEMIS: (i) outperforms state-of-art methods to predict held-out timepoints, (ii) recovers relative cell population changes over time, and (iii) identifies "drift" genes driving deterministic expression trends in cell trajectories. Furthermore, in silico perturbations show that these genes influence processes like EMT.
Availability and implementation: The code for ARTEMIS: https://github.com/daifengwanglab/ARTEMIS.
© The Author(s) 2025. Published by Oxford University Press.
Figures




Update of
-
ARTEMIS integrates autoencoders and Schrödinger Bridges to predict continuous dynamics of gene expression, cell population and perturbation from time-series single-cell data.bioRxiv [Preprint]. 2025 Jan 26:2025.01.23.634618. doi: 10.1101/2025.01.23.634618. bioRxiv. 2025. Update in: Bioinformatics. 2025 Jul 1;41(Supplement_1):i189-i197. doi: 10.1093/bioinformatics/btaf218. PMID: 39896674 Free PMC article. Updated. Preprint.
References
-
- Bernton E, Heng J, Doucet A et al. Schrödinger bridge samplers. arXiv, arXiv:1912.13170, 2019, preprint: not peer reviewed.
-
- Chen T, Liu G-H, Theodorou EA. Likelihood training of schrödinger bridge using forward-backward SDES theory. In: International Conference on Learning Representations. Virtual Event, 2022.
-
- Cuturi M. Sinkhorn distances: lightspeed computation of optimal transport. Adv Neural Inf Process Syst 2013;26:2292–300.
-
- De Bortoli V, Thornton J, Heng J et al. Diffusion Schrödinger bridge with applications to score-based generative modeling. Adv Neural Inf Process Syst 2021;34:17695–709.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

