Monoallelic mutations in MMD2 cause autosomal dominant aggressive periodontitis
- PMID: 40663042
- PMCID: PMC12262042
- DOI: 10.1084/jem.20231911
Monoallelic mutations in MMD2 cause autosomal dominant aggressive periodontitis
Abstract
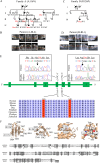
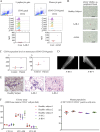
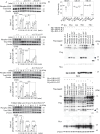
Aggressive periodontitis causes rapid destruction of periodontal tissue. It occurs at a young age with familial clustering. We report on the first time on molecular and cellular basis of a Mendelian form of autosomal dominant aggressive periodontitis. Monoallelic mutations in the monocyte to macrophage differentiation-associated 2 (MMD2) gene, encoding MMD2, in two Japanese families with autosomal dominant aggressive periodontitis are identified. Mutations, c.347 C>T (p.A116V) and c.377 G>C (p.R126P) in MMD2, disturbed fMLP-induced activation of Ras/ERK signaling. Additionally, abnormalities in the proteins of Golgi apparatus, a crucial contributor to innate immune signaling pathways, were identified in patients' neutrophils. The knock-in and knockout mice exhibited alveolar bone loss by ligature-induced periodontitis, along with impaired fMLP-induced chemotaxis, as found in the patients with MMD2 mutation. Our studies revealed that monoallelic mutations in MMD2 underlie the impairment of neutrophil chemotaxis, which leads to the development of autosomal dominant aggressive periodontitis.
© 2025 Iwata et al.
Conflict of interest statement
Disclosures: H. Kanegane reported personal fees from Takeda Pharmaceutical Co. outside the submitted work. S. Okada reported grants from Japan Agency for Medical Research and Development, and from the Japan Society for the Promotion of Science during the conduct of the study. No other disclosures were reported.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

