Nodal modulator (NOMO) is a force-bearing transmembrane protein required for muscle differentiation
- PMID: 40663102
- PMCID: PMC12262048
- DOI: 10.1083/jcb.202505010
Nodal modulator (NOMO) is a force-bearing transmembrane protein required for muscle differentiation
Abstract
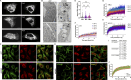
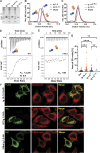
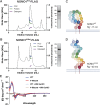
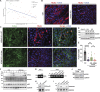
The ER relies on membrane-shaping proteins to maintain a continuous network of sheets and tubules that host distinct biological processes. How this intricate structure of the ER membrane system is maintained under conditions of mechanical strain is incompletely understood. NOMO is an ER-resident transmembrane protein that contributes to ER morphology and is highly expressed in striated muscle. In this study, we identify a critical interface between distal Ig domains that enables NOMO to maintain ER morphology and bear mechanical forces. By incorporating two independent tension sensors in the luminal domain of NOMO, we demonstrate that NOMO assemblies experience forces in the single piconewton range, with a significant contribution from the identified interface. These newly defined features are important-if not indispensable-for myogenesis, as interface mutations affecting mechanosensitivity fail to restore the essential role of NOMO during myogenesis in a C2C12 differentiation model. Moreover, NOMO depletion impairs nematode motility, underscoring a broader functional importance in muscle physiology.
© 2025 Naughton et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. J. Berro reported a patent to PCT/US2023/069505 pending. No other disclosures were reported.
Figures









Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Adenovirus Modulates Toll-Like Receptor 4 Signaling by Reprogramming ORP1L-VAP Protein Contacts for Cholesterol Transport from Endosomes to the Endoplasmic Reticulum.J Virol. 2017 Feb 28;91(6):e01904-16. doi: 10.1128/JVI.01904-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077646 Free PMC article.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Interventions for promoting habitual exercise in people living with and beyond cancer.Cochrane Database Syst Rev. 2018 Sep 19;9(9):CD010192. doi: 10.1002/14651858.CD010192.pub3. Cochrane Database Syst Rev. 2018. PMID: 30229557 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
Genetically encoded mechano-sensors with versatile readouts and compact size.bioRxiv [Preprint]. 2025 Jul 1:2025.01.16.633409. doi: 10.1101/2025.01.16.633409. bioRxiv. 2025. PMID: 39868242 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

