Loss of IL13RA2 promotes metastatic tumor growth in triple-negative breast cancer via increased AKT and NF-κB signaling
- PMID: 40663259
- PMCID: PMC12263738
- DOI: 10.1007/s10585-025-10362-1
Loss of IL13RA2 promotes metastatic tumor growth in triple-negative breast cancer via increased AKT and NF-κB signaling
Abstract
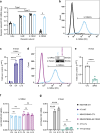
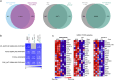
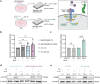
Triple-negative breast cancer is associated with poor patient prognosis and high rates of distant metastasis. These patients are at elevated risk of brain metastasis, which remains a major therapeutic challenge. IL13RA2, a high-affinity receptor for IL13, is highly expressed in primary brain cancers, many extracranial solid tumors, and in lung- and brain-seeking metastatic variant cell lines. However, the relationship between IL13RA2 and patient prognosis is variable, and the biological function of this receptor in cancer remains controversial. We sought to define the role of IL13RA2 in triple-negative breast cancer growth and metastasis, with an emphasis on breast-to-brain metastasis. We generated IL13RA2-CRISPR knockout derivatives of the human brain-seeking breast cancer cell line MDA231BrM2, as well as murine 4T1 cells, and evaluated changes in gene expression, proliferation, survival, and metastatic growth in vivo. Both IL13RA2-deficient models demonstrate enhanced cell survival in vitro, as well as augmented metastatic tumor growth and worsened animal survival in intracardiac models of brain metastasis. Concordantly, elevated IL13RA2 mRNA expression is positively correlated with overall survival in patients with basal-like breast cancer. Mechanistically, IL13RA2-deficient cells exhibit increased AKT and NF-κB signaling. These cells are sensitive to inhibition of either pathway, but especially AKT, which may represent a clinically useful vulnerability for patients with IL13RA2-low tumors. Our data suggest that inhibition of IL13RA2, though promising in other tumor contexts, may be deleterious in metastatic triple-negative breast cancer.
Keywords: AKT; Brain metastasis; IL13RA2; NF-κB; Signaling; Triple-negative breast cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: All animal studies were approved by Vanderbilt’s Institutional Animal Care and Use Committee (protocol M2300038-00). Competing interests: Barbara Fingleton is a member of the editorial board for Clinical and Experimental Metastasis.
Figures



Similar articles
-
Inhibition of Interleukin-8/C-X-C Chemokine Receptor 2 Signaling Axis Prevents Tumor Growth and Metastasis in Triple-Negative Breast Cancer Cells.Pharmacology. 2025;110(3):178-190. doi: 10.1159/000545659. Epub 2025 Apr 4. Pharmacology. 2025. PMID: 40188812 Free PMC article.
-
H3K36me2 methyltransferase NSD2/WHSC1 promotes triple-negative breast cancer metastasis via activation of ULK1-dependent autophagy.Autophagy. 2025 Aug;21(8):1824-1842. doi: 10.1080/15548627.2025.2479995. Epub 2025 Mar 25. Autophagy. 2025. PMID: 40097917 Free PMC article.
-
α9 Nicotinic Acetylcholine Receptor Promotes Tumor Proliferation and Suppresses Ferroptosis in Triple-Negative Breast Cancer.Biomolecules. 2025 Jun 8;15(6):835. doi: 10.3390/biom15060835. Biomolecules. 2025. PMID: 40563476 Free PMC article.
-
Platinum-containing regimens for metastatic breast cancer.Cochrane Database Syst Rev. 2017 Jun 23;6(6):CD003374. doi: 10.1002/14651858.CD003374.pub4. Cochrane Database Syst Rev. 2017. PMID: 28643430 Free PMC article.
-
An update on cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer.Future Oncol. 2025 Mar;21(6):715-735. doi: 10.1080/14796694.2025.2461443. Epub 2025 Feb 12. Future Oncol. 2025. PMID: 39936282 Review.
References
-
- Cancer Stat Facts: Female Breast Cancer Subtypes (SEER 22 2017–2021) National Cancer institute. https://www.seer.cancer.gov. Accessed 5 December 2024
-
- Triple Negative Breast Cancer (2021) PathologyOutlines.com. https://www.pathologyoutlines.com/topic/breastmalignanttripleneg.html. Accessed 15 February 2025
-
- Costa RLB, Gradishar WJ (2017) Triple-negative breast cancer: current practice and future directions. J Oncol Pract 13(5):301–303. 10.1200/jop.2017.023333 - PubMed
-
- Ostrom QT, Wright CH, Barnholtz-Sloan JS (2018) Chap. 2: brain metastases: epidemiology. In: Schiff D, Van den Bent MJ (eds) Handbook of clinical neurology. Elsevier, pp 27–42 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

