Bayesian network imputation methods applied to multi-omics data identify putative causal relationships in a type 2 diabetes dataset containing incomplete data: An IMI DIRECT Study
- PMID: 40663565
- PMCID: PMC12279144
- DOI: 10.1371/journal.pgen.1011776
Bayesian network imputation methods applied to multi-omics data identify putative causal relationships in a type 2 diabetes dataset containing incomplete data: An IMI DIRECT Study
Abstract
Here we report the results from exploratory analysis using a Bayesian network approach of data originally derived from a large North European study of type 2 diabetes (T2D) conducted by the IMI DIRECT consortium. 3029 individuals (795 with T2D and 2234 without) within 7 different study centres provided data comprising genotypes, proteins, metabolites, gene expression measurements and many different clinical variables. The main aim of the current study was to demonstrate the utility of our previously developed method to fit Bayesian networks by performing exploratory analysis of this dataset to identify possible causal relationships between these variables. The data was analysed using the BayesNetty software package, which can handle mixed discrete/continuous data with missing values. The original dataset consisted of over 16,000 variables, which were filtered down to 260 variables for analysis. Even with this reduction, no individual had complete data for all variables, making it impossible to analyse using standard Bayesian network methodology. However, using the recently proposed novel imputation method implemented in BayesNetty we computed a large average Bayesian network from which we could infer possible associations and causal relationships between variables of interest. Our results confirmed many previous findings in connection with T2D, including possible mediating proteins and genes, some of which have not been widely reported. We also confirmed potential causal relationships with liver fat that were identified in an earlier study that used the IMI DIRECT dataset but was limited to a smaller subset of individuals and variables (namely individuals with complete data at pre-defined variables of interest). In addition to providing valuable confirmation, our analyses thus demonstrate a proof-of-principle of the utility of the method implemented within BayesNetty. The full final average Bayesian network generated from our analysis is freely available and can be easily interrogated further to address specific focussed scientific questions of interest.
Copyright: © 2025 Howey et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: HJC and AV serve on the editorial board of PLOS Genetics. MMcC and AM are currently employees of Genentech and holders of Roche stock.
Figures

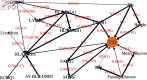
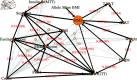


References
-
- Koivula RW, Heggie A, Barnett A, Cederberg H, Hansen TH, Koopman AD, et al. Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: rationale and design of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia. 2014;57(6):1132–42. doi: 10.1007/s00125-014-3216-x - DOI - PMC - PubMed
-
- Koivula RW, Forgie IM, Kurbasic A, Viñuela A, Heggie A, Giordano GN, et al. Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: descriptive characteristics of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia. 2019;62(9):1601–15. doi: 10.1007/s00125-019-4906-1 - DOI - PMC - PubMed
-
- Atabaki NN, Coral DE, Pomares-Millan H, Smith K, Behjat HH, Koivula RW, et al. A biological-systems-based analyses using proteomic and metabolic network inference reveals mechanistic insights into hepatic lipid accumulation: an IMI-DIRECT study. medRxiv. 2025:2025.06.02.25328773. doi: 10.1101/2025.06.02.25328773 - DOI - PMC - PubMed
-
- Howey R, Shin S-Y, Relton C, Davey Smith G, Cordell HJ. Bayesian network analysis incorporating genetic anchors complements conventional Mendelian randomization approaches for exploratory analysis of causal relationships in complex data. PLoS Genet. 2020;16(3):e1008198. doi: 10.1371/journal.pgen.1008198 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

