Variable lymphocyte receptor F is generated via somatic diversification and expressed by lamprey T-like cells
- PMID: 40664669
- PMCID: PMC12264053
- DOI: 10.1038/s41467-025-61187-1
Variable lymphocyte receptor F is generated via somatic diversification and expressed by lamprey T-like cells
Abstract
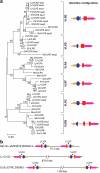
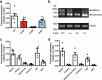
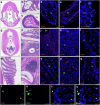
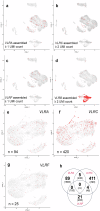
All extant jawless vertebrates (lampreys and hagfishes) possess a unique adaptive immune system characterized by highly variable lymphocyte receptors (VLR) that are assembled in developing lymphocytes using leucine-rich-repeat donor cassettes. Five VLR types have been identified in lampreys: VLRA, VLRB, VLRC, VLRD, and VLRE. VLRB-expressing lymphocytes are functional analogs to B cells, whereas VLRA, VLRC, VLRD, and VLRE-expressing lymphocytes are more akin to T cells of jawed vertebrates. Here we define an additional VLR, designated VLRF. VLRF is phylogenetically closest to VLRA, with which it likely shares a common ancestral gene of at least 250 million years in the past. VLR assembly analyses show that VLRA, VLRC, VLRD, VLRE, and VLRF share donor cassettes through long-range intra- and inter-chromosomal interactions, whereas VLRB utilizes a distinct, dedicated cassette set. The pattern of gene expression, donor cassette usage, and distinctive amino acid composition in the C-terminal stalk suggest that VLRF⁺ lymphocytes may represent an additional T-like sub-lineage, adding further complexity to the VLR-based adaptive immune system.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Evolution of two distinct variable lymphocyte receptors in lampreys: VLRD and VLRE.Cell Rep. 2023 Aug 29;42(8):112933. doi: 10.1016/j.celrep.2023.112933. Epub 2023 Aug 4. Cell Rep. 2023. PMID: 37542721 Free PMC article.
-
Genomic donor cassette sharing during VLRA and VLRC assembly in jawless vertebrates.Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14828-33. doi: 10.1073/pnas.1415580111. Epub 2014 Sep 16. Proc Natl Acad Sci U S A. 2014. PMID: 25228758 Free PMC article.
-
Evolution of variable lymphocyte receptor B antibody loci in jawless vertebrates.Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2116522118. doi: 10.1073/pnas.2116522118. Proc Natl Acad Sci U S A. 2021. PMID: 34880135 Free PMC article.
-
Evolution of adaptive immunity: implications of a third lymphocyte lineage in lampreys.Bioessays. 2014 Mar;36(3):244-50. doi: 10.1002/bies.201300145. Epub 2013 Dec 19. Bioessays. 2014. PMID: 24853392 Review.
-
Leveraging the biotechnological promise of the hagfish variable lymphocyte receptors: tools for aquatic microbial diseases.Fish Shellfish Immunol. 2024 Jul;150:109565. doi: 10.1016/j.fsi.2024.109565. Epub 2024 Apr 17. Fish Shellfish Immunol. 2024. PMID: 38636740 Review.
References
-
- Holland, N. D. & Chen, J. Origin and early evolution of the vertebrates: new insights from advances in molecular biology, anatomy, and palaeontology. Bioessays23, 142–151 (2001). - PubMed
-
- Janvier, P. Early jawless vertebrates and cyclostome origins. Zool. Sci.25, 1045–1056 (2008). - PubMed
-
- Kuratani, S. Evo-devo studies of cyclostomes and the origin and evolution of jawed vertebrates. Curr. Top. Dev. Biol.141, 207–239 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

