Secondary metabolite biosynthetic gene clusters and microbial diversity as predicted by shotgun metagenomic sequencing approach in hospitals and pharmaceutical industry wastes
- PMID: 40664768
- PMCID: PMC12264290
- DOI: 10.1038/s41598-025-03467-w
Secondary metabolite biosynthetic gene clusters and microbial diversity as predicted by shotgun metagenomic sequencing approach in hospitals and pharmaceutical industry wastes
Abstract
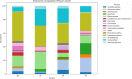
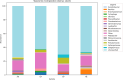
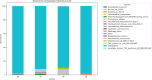
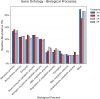
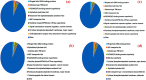
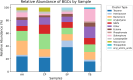
Hospital and pharmaceutical industry wastes harbor a diverse microbial community influenced by their complex chemical composition, including antibiotic-rich compounds originating from soil microorganisms. This study employs a shotgun metagenomic approach to comprehensively characterize the microbial composition of hospital and Pharmaceutical industry wastes for the first time in Ethiopia. Metagenomic DNA was extracted and used to construct a whole-genome shotgun library, which was subsequently sequenced using the Illumina HiSeq1500 platform. Taxonomic analysis revealed that bacteria were the predominant domain across all samples, followed by eukaryotes and archaea. Among the bacterial phyla, Pseudomonadota was the most prevalent in both hospital and pharmaceutical waste samples. At the genus level, Pseudomonas and Pedobacter were the most abundant taxa, followed by Flavobacterium and Streptomyces. Notably, Streptomyces exhibited higher-than-expected abundance in the waste metagenome, suggesting a potential adaptive response to environmental stressors. Across all samples, Pedobacter and Pseudomonas were consistently the most dominant bacterial genera. Functional analysis using gene ontology (GO) annotations highlighted the predominance of metabolic and biosynthetic processes. Further investigation revealed an enrichment of ATP-binding cassette (ABC) transporter protein families and winged-helix protein domains, both of which are linked to antibiotic resistance, metabolite translocation, and antibiotic biosynthesis regulation. KEGG pathway analysis identified key biosynthetic pathways, including terpenoid and polyketide biosynthesis, as well as beta-lactam antibiotic production. Additionally, antiSMASH analysis detected multiple biosynthetic gene clusters (BGCs), including those encoding terpenes, bacteriocins, and non-ribosomal peptide synthetases. Overall, this study underscores the adaptive potential of microorganisms inhabiting industrial waste environments, highlighting their genomic capacity to produce bioactive secondary metabolites in response to toxic compounds. These findings provide valuable insights into microbial resilience and the potential for biotechnological applications in bioremediation and drug discovery.
Keywords: Biosynthesis gene clusters; Hospital wastes; Metagenomics; Microbial diversity; Pharmaceutical wastes; Shotgun sequencing; Unculturable bacteria.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: Not applicable. Consent for publication: Not applicable.
Figures

Similar articles
-
Novel Gene Clusters for Natural Product Synthesis Are Abundant in the Mangrove Swamp Microbiome.Appl Environ Microbiol. 2023 Jun 28;89(6):e0010223. doi: 10.1128/aem.00102-23. Epub 2023 May 16. Appl Environ Microbiol. 2023. PMID: 37191511 Free PMC article.
-
Genome mining identifies a diversity of natural product biosynthetic capacity in human respiratory Corynebacterium strains.mSphere. 2025 Jun 25;10(6):e0025825. doi: 10.1128/msphere.00258-25. Epub 2025 May 21. mSphere. 2025. PMID: 40396729 Free PMC article.
-
Genome mining reveals the distribution of biosynthetic gene clusters in Alternaria and related fungal taxa within the family Pleosporaceae.BMC Genomics. 2025 Jul 21;26(1):678. doi: 10.1186/s12864-025-11754-z. BMC Genomics. 2025. PMID: 40691532 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Interventions to improve antibiotic prescribing practices for hospital inpatients.Cochrane Database Syst Rev. 2017 Feb 9;2(2):CD003543. doi: 10.1002/14651858.CD003543.pub4. Cochrane Database Syst Rev. 2017. PMID: 28178770 Free PMC article.
References
-
- Tahrani, L. et al. Isolation and characterization of antibiotic-resistant bacteria from pharmaceutical industrial wastewaters. Microb. Pathog. 89, 54–61 (2015). - PubMed
-
- Hauhnar, L., Pachuau, L. & Lalhruaitluanga, H. Isolation and characterization of Multi-Drug resistant Bacteria from hospital wastewater sites around the City of Aizawl, Mizoram. Adv. Bioscience Biotechnol.9 (07), 311–321 (2018).
-
- WHO. Lack of New Antibiotics Threatens Global Efforts to Contain Drug-Resistant Infections. (2020).
MeSH terms
LinkOut - more resources
Full Text Sources

