Germline genetic variation impacts clonal hematopoiesis landscape and progression to malignancy
- PMID: 40664769
- PMCID: PMC12339400
- DOI: 10.1038/s41588-025-02250-x
Germline genetic variation impacts clonal hematopoiesis landscape and progression to malignancy
Abstract
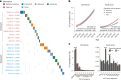
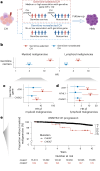
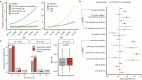
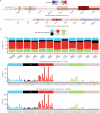
With age, clonal expansions occur pervasively across normal tissues yet only in rare instances lead to cancer, despite being driven by well-established cancer drivers. Characterization of the factors that influence clonal progression is needed to inform interventional approaches. Germline genetic variation influences cancer risk and shapes tumor mutational profile, but its influence on the mutational landscape of normal tissues is not well known. Here we studied the impact of germline genetic variation on clonal hematopoiesis (CH) in 731,835 individuals. We identified 22 new CH-predisposition genes, most of which predispose to CH driven by specific mutational events. CH-predisposition genes contribute to unique somatic landscapes, reflecting the influence of germline genetic backdrop on gene-specific CH fitness. Correspondingly, somatic-germline interactions influence the risk of CH progression to hematologic malignancies. These results demonstrate that germline genetic variation influences somatic evolution in the blood, findings that likely extend to other tissues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.L.B. reports research grants and personal fees from Servier. E.P. is the co-founder of and holds a fiduciary role in Isabl Inc. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech or Roche and Novartis; personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Bristol Myers Squibb, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Capital, Foresite Labs, Genentech or Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, Novo Nordisk, TenSixteen Bio and Tourmaline Bio; equity in Bolt, Candela, Mercury, MyOme, Parameter Health, Preciseli and TenSixteen Bio; and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. R.L.L. is on the Supervisory Board of QIAGEN (compensation or equity), a co-founder of or board member at Ajax (equity) and a scientific advisor to Mission Bio, Kurome, Anovia, Bakx, Syndax, Scorpion, Zentalis, Auron, Prelude and C4 Therapeutics; for each of these entities he receives equity or compensation. He has received research support from the Cure Breast Cancer Foundation, Calico, Zentalis and Ajax, and has consulted for Jubilant, Goldman Sachs, Incyte, AstraZeneca and Janssen. A.G.B. reports personal fees and equity in TenSixteen Bio, all unrelated to this work. The other authors declare no competing interests.
Figures




References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

