Cold exposure promotes the progression of osteoarthritis through downregulating APOE in cartilage
- PMID: 40664892
- PMCID: PMC12340072
- DOI: 10.1038/s44321-025-00268-6
Cold exposure promotes the progression of osteoarthritis through downregulating APOE in cartilage
Abstract
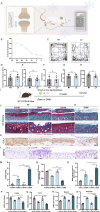
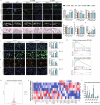
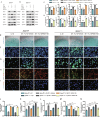
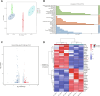
Osteoarthritis (OA) is a degenerative joint disease with limited effective therapies. Cold weathers have been shown to affect joint pain in OA patients. However, the impact of cold climate on OA progression is debated, with the underlying mechanisms not fully understood. This study aims to elucidate the role of Apolipoprotein E (Apoe) in chondrocytes in relation to OA progression under cold exposure. Both human chondrocytes RNA sequencing and DMM mice OA model revealed that lower temperatures significantly downregulated Apoe expression, correlating with OA exacerbation. Conditional knockout of Apoe in cartilage aggravated cartilage degeneration, leading to lipid accumulation, increased ROS production, mitochondrial dysfunction, and elevated chondrocyte apoptosis. Treatment with RGX-104, an LXRβ agonist, reversely restored APOE expression, mitigated aberrant lipid accumulation and countered the detrimental effects of cold exposure on OA progression. These results suggest that targeting lipid transfer and metabolism, especially through Apoe modulation, may offer therapeutic strategies for OA patients residing in colder climates, such as those at high altitudes and latitudes, and even winter season.
Keywords: Apolipoprotein E; Cold Exposure; Lipid Accumulation; Osteoarthritis; ROS.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures





References
-
- Bonner JrWM, Jonsson JrH, Malanos C, Bryant M (1975) Changes in the lipids of human articular cartilage with age. Arthritis Rheumatism 18:461–473 - PubMed
-
- Choi W-S, Lee G, Song W-H, Koh J-T, Yang J, Kwak J-S, Kim H-E, Kim SK, Son Y-O, Nam H et al (2019) The CH25H–CYP7B1–RORα axis of cholesterol metabolism regulates osteoarthritis. Nature 566:254–258 - PubMed
MeSH terms
Substances
Grants and funding
- 21YF1405800/Shanghai sailing program
- PW2024B-07/Shanghai Pudong New Area Health Commission ( )
- PKJ2020-Y44/Shanghai Pudong New Area Health Commission ( )
- Pwyts2021-03/The Featured Clinical Discipline Project of Shanghai Pudong New District
- ZR202102210485/Shandong Provincial Natural Science Foundation
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

