Fibroblast growth factor receptor signaling modulates cholesterol storage in a SOAT1-dependent manner to promote mammary tumor cell invasion
- PMID: 40665359
- PMCID: PMC12261779
- DOI: 10.1186/s13058-025-02084-9
Fibroblast growth factor receptor signaling modulates cholesterol storage in a SOAT1-dependent manner to promote mammary tumor cell invasion
Abstract
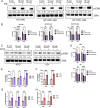
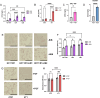
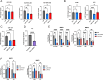
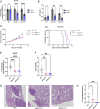
Signaling by fibroblast growth factor receptors (FGFRs) is active in up to 85% of breast cancers and results in enhanced proliferation, migration, and invasion of tumor cells. Here, we show that FGFR signaling regulates cholesterol metabolism in breast cancer. Specifically, we demonstrate that FGFR activation promotes cellular cholesterol storage by upregulating expression of the enzyme sterol O-acyltransferase 1 (SOAT1). Moreover, we demonstrate that inhibition of SOAT1 attenuates FGFR-driven colony formation and invasion in tumor cells, which correlates with reduced expression of matrix metalloproteinase expression. Furthermore, genetic knockdown of SOAT1 decreases mammary tumor growth in vivo. Taken together, these findings suggest a largely undiscovered metabolic role for FGFR signaling in regulating cholesterol metabolism in breast cancer and present a therapeutic vulnerability that could be targeted in FGFR-driven cancers.
Keywords: Cholesterol metabolism; Cholesterol storage; Fibroblast growth factor; Triple-negative breast cancer.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal care and procedures were approved by the University of Minnesota IACUC under protocol 2208–40316 A and were in accordance with the procedures detailed in the Guide for the Care and Use of Laboratory Animals. No human subjects were used in this study. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16(2):179–86. - PubMed
-
- Courjal F, Cuny M, Simony-Lafontaine J, Louason G, Speiser P, Zeillinger R, et al. Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups. Cancer Res. 1997;57(19):4360–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

