The dynamics of the gut microbiota in prediabetes during a four-year follow-up among European patients-an IMI-DIRECT prospective study
- PMID: 40665409
- PMCID: PMC12261804
- DOI: 10.1186/s13073-025-01508-7
The dynamics of the gut microbiota in prediabetes during a four-year follow-up among European patients-an IMI-DIRECT prospective study
Abstract
Background: Previous case-control studies have reported aberrations of the gut microbiota in individuals with prediabetes. The primary objective of the present study was to explore the dynamics of the gut microbiota of individuals with prediabetes over 4 years with a secondary aim of relating microbiota dynamics to temporal changes of metabolic phenotypes.
Methods: The study included 486 European patients with prediabetes. Gut microbiota profiling was conducted using shotgun metagenomic sequencing and the same bioinformatics pipelines at study baseline and after 4 years. The same phenotyping protocols and core laboratory analyses were applied at the two timepoints. Phenotyping included anthropometrics and measurement of fasting plasma glucose and insulin levels, mean plasma glucose and insulin under an oral glucose tolerance test (OGTT), 2-h plasma glucose after an OGTT, oral glucose insulin sensitivity index, Matsuda insulin sensitivity index, body mass index, waist circumference, and systolic and diastolic blood pressure. Measures of the dynamics of bacterial microbiota were related to concomitant changes in markers of host metabolism.
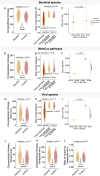
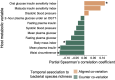
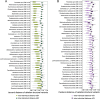
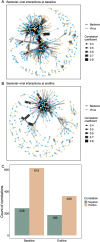
Results: Over 4 years, significant declines in richness were observed in gut bacterial and viral species and microbial pathways accompanied by significant changes in the relative abundance and the genetic composition of multiple bacterial species. Additionally, bacterial-viral interactions diminished over time. Despite the overall reduction in bacterial richness and microbial pathway richness, 80 dominant core bacterial species and 78 core microbial pathways were identified at both timepoints in 99% of the individuals, representing a resilient component of the gut microbiota. Over the same period, individuals with prediabetes exhibited a significant increase in glycemia and insulinemia alongside a significant decline in insulin sensitivity. Estimates of the gut bacterial microbiota dynamics were significantly correlated with temporal impairments in host metabolic health.
Conclusions: In this 4-year prospective study of European patients with prediabetes, the gut microbiota exhibited major changes in taxonomic composition, bacterial species genetics, and microbial functional potentials, many of which paralleled an aggravation of host metabolism. Whether the temporal gut microbiota changes represent an adaptation to the progression of metabolic abnormalities or actively contribute to these in prediabetes cases remains unsettled.
Trial registration: The Diabetes Research on Patient Stratification (DIRECT) study, an exploratory observational study initiated on October 15, 2012, was registered on ClinicalTrials.gov under the number NCT03814915.
Keywords: Gut bacterial genetics; Gut bacterial microbiota; Gut viral microbiota; Insulin sensitivity; Long-term dynamics; Metabolism; Microbial functional pathways; Prediabetes.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate.: The DIRECT study (Diabetes Research on Patient Stratification) was registered under ClinicalTrials.gov ID NCT03814915. Approval of the study protocol was obtained from each of the regional research ethics review boards separately: Lund, Sweden: 20130312105459927; Copenhagen, Denmark: H-1–2012-166 and H-1–2012-100; Amsterdam, the Netherlands: NL40099.029.12; Newcastle, Dundee, and Exeter, UK: 12/NE/0132. All participants gave written informed consent to participate in the study at enrollment and the research conformed to the ethical principles for medical research involving human participants outlined in the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: RK has received consulting fees from Novo Nordisk; he was also funded by a STAR Award Novo Nordisk co-financed PhD fellowship and a Novo Nordisk Foundation postdoctoral fellowship (NNF18OC0031650). PWF has received research funding from Boehringer Ingelheim, Eli Lilly, Janssen, Novo Nordisk A/S, Sanofi Aventis and Servier, received consulting fees from Eli Lilly, Novo Nordisk and Zoe Global Ltd and has stock options in Zoe Global Ltd. HR is an employee of Boehringer Ingelheim and a shareholder of Sanofi Aventis. MMcC declares that the views expressed in this article are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health; he has served on advisory panels for Pfizer, Novo Nordisk and Zoe Global, has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly, and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda; as of June 2019, he is an employee of Genentech, and a holder of Roche stock. BJ and PBM are employees of Sanofi Deutschland GmbH. IP is employed by Eli Lilly Regional Operations GmbH. HR is an employee of Boehringer Ingelheim International GmbH. MR is employed by Novo Nordisk A/S. OP and YF are co-founders of GutCRINE. The remaining authors declare that they have no competing interests.
Figures



References
-
- American Diabetes Association. 2. Classification and diagnosis of diabetes. Diabetes Care. 2016;40:S11-24. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

