A Kinetic Model of Antigen-Dependent IgG Oligomerization and Complement Binding
- PMID: 40666036
- PMCID: PMC12257888
- DOI: 10.1002/smsc.202500149
A Kinetic Model of Antigen-Dependent IgG Oligomerization and Complement Binding
Abstract
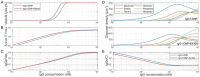
The classical complement pathway (CCP) is an essential part of the immune system, activated when complement protein C1 binds to IgG antibody oligomers on the surface of pathogens, infected or malignant cells, culminating in the formation of the membrane attack complex and subsequent cell lysis. IgG oligomers also engage immune effector cells through Fcγ receptors or complement receptors, facilitating antibody-dependent cellular cytotoxicity and phagocytosis. Understanding the factors that drive IgG oligomerization is thus crucial for improving IgG-based therapies. Herein, a kinetic model to predict oligomer formation based on IgG concentration, antigen density, IgG subclass, Fc mutants, and oligomerization inhibitors like staphylococcal protein A is developed. The underlying molecular interactions in single molecule force spectroscopy and grating coupled interferometry experiments are characterized. By fitting experimental data from high-speed atomic force microscopy experiments, key rate constants and thermodynamic parameters, including free energy changes associated with oligomerization and apply the model to predict complement-mediated lysis in liposomal vesicle-based assays, are further quantified. The presented mechanistic framework may serve as a basis for optimizing antibody engineering and pharmacokinetic/pharmacodynamic modeling in the context of immunotherapies exploiting the CCP.
Keywords: C1; IgG hexamers; IgG oligomerization; IgG subclasses; classical complement pathway; complement mediated; kinetic oligomerization model; lysis.
© 2025 The Author(s). Small Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare the following competing financial interest(s): F.J.B., S.B., and A.F.L., are inventors on patent applications related to complement activation by therapeutic antibodies and own Genmab stock. J.P. received Genmab funding.
Figures






References
-
- Ugurlar D., Howes S. C., de Kreuk B.‐J., Koning R. I., de Jong R. N., Beurskens F. J., Schuurman J., Koster A. J., Sharp T. H., Parren P. W. H. I., Gros P., Science 2018, 359, 794. - PubMed
-
- Strasser J., de Jong R. N., Beurskens F. J., Wang G., Heck A. J. R., Schuurman J., Parren P. W. H. I., Hinterdorfer P., Preiner J., Nano Lett. 2019, 19, 4787. - PubMed
-
- Strasser J., de Jong R. N., Beurskens F. J., Schuurman J., Parren P. W. H. I., Hinterdorfer P., Preiner J., ACS Nano 2020, 14, 2739. - PubMed
LinkOut - more resources
Full Text Sources
