Surveillance of coronaviruses in wild aquatic birds in Hong Kong: expanded genetic diversity and discovery of novel subgenus in the Deltacoronavirus
- PMID: 40666308
- PMCID: PMC12262178
- DOI: 10.1093/ve/veaf049
Surveillance of coronaviruses in wild aquatic birds in Hong Kong: expanded genetic diversity and discovery of novel subgenus in the Deltacoronavirus
Abstract
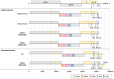
Migratory birds may carry emerging viruses over long distances. Regular surveillance and metagenomic analysis were employed to explore the diversity of avian coronaviruses at Hong Kong's Mai Po Wetland. We tested a total of 3239 samples collected from 2018 to 2024, among which the prevalence rate of viruses of the genus Gammacoronavirus (64.4%) was higher than that of Deltacoronavirus (35.6%). The host species were identified for 79.8% of the coronavirus-positive samples. Two deltacoronaviruses with full-genome sequences and one nearly complete gammacoronavirus genome were identified in faecal samples of three bird species. We also predicted putative transcriptional regulatory sequences and 3CLpro and PLpro cleavage sites for these viruses. Results from our phylogenetic analysis and pairwise amino acid identity comparisons, using the International Committee on Taxonomy of Viruses classification criteria based on the DEmARC framework, indicate that black-faced spoonbill coronavirus (BSCoV, strain MP22-1474) prototypes a new subgenus. Great cormorant coronavirus (GCCoV, strain MP18-1070) and falcated duck coronavirus (FDCoV, strain MP22-196) belong to two previously known species while diverging most profoundly from known viruses of these species. Two recombination events may have contributed to the evolution of FDCoV MP22-196 in genome regions from ORF1b to the S gene and from the M gene to the N gene. The cophylogenetic analysis between avian hosts and coronaviruses provides evidence for a strong linkage between viruses of the genus Gammacoronavirus and the birds of order Anseriformes. This study highlights the importance of ongoing surveillance for coronaviruses in wild migratory birds.
Keywords: Deltacoronavirus; Gammacoronavirus; subgenus; virus surveillance; wild aquatic birds.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures








Similar articles
-
Long-term maintenance of a Deltacoronavirus infecting multiple bird species in Antarctica.Microbiol Spectr. 2025 Aug 5;13(8):e0268824. doi: 10.1128/spectrum.02688-24. Epub 2025 Jun 16. Microbiol Spectr. 2025. PMID: 40522158 Free PMC article.
-
Discovery, phylogenetic, and comparative genomic analysis of novel avian gammacoronaviruses identified in feral pigeons (Columba livia domestica).J Virol. 2025 Aug 20:e0111225. doi: 10.1128/jvi.01112-25. Online ahead of print. J Virol. 2025. PMID: 40833823
-
Genetic and Molecular Characterization of Avian Influenza A(H9N2) Viruses from Live Bird Markets (LBM) in Senegal.Viruses. 2025 Jan 8;17(1):73. doi: 10.3390/v17010073. Viruses. 2025. PMID: 39861862 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Nicotine receptor partial agonists for smoking cessation.Cochrane Database Syst Rev. 2016 May 9;2016(5):CD006103. doi: 10.1002/14651858.CD006103.pub7. Cochrane Database Syst Rev. 2016. Update in: Cochrane Database Syst Rev. 2023 May 5;5:CD006103. doi: 10.1002/14651858.CD006103.pub8. PMID: 27158893 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources

