This is a preprint.
Genomic characterization of normal and aberrant human milk production
- PMID: 40666332
- PMCID: PMC12262777
- DOI: 10.1101/2025.06.22.25329156
Genomic characterization of normal and aberrant human milk production
Update in
-
Genomic characterization of normal and aberrant human milk production.Sci Adv. 2025 Sep 12;11(37):eadr7174. doi: 10.1126/sciadv.adr7174. Epub 2025 Sep 10. Sci Adv. 2025. PMID: 40929253 Free PMC article.
Abstract
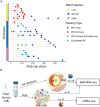
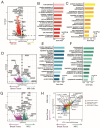
Breastfeeding is essential for reducing infant morbidity and mortality, yet exclusive breastfeeding rates remain low, often due to insufficient milk supply. The molecular causes of low milk production are not well understood. Fresh milk samples from 30 lactating individuals, classified by milk production levels across postpartum stages, were analyzed using genomic and microbiome techniques. Bulk RNA sequencing of milk fat globules (MFG), milk cells, and breast tissue revealed that MFG-derived RNA closely mirrors luminal milk cells. Transcriptomic and single-cell RNA analyses identified changes in gene expression and cellular composition, highlighting key genes (GLP1R, PLIN4, KLF10) and cell-type differences between low and high producers. Infant microbiome diversity was influenced by feeding type but not maternal milk supply. This study provides a comprehensive human milk transcriptomic catalog and highlights that MFG could serve as a useful biomarker for milk transcriptome analysis, offering insights into the genetic factors influencing milk production.
Conflict of interest statement
Competing interests: BEE is on the Scientific Advisory Board for ArrePath Inc, Crayon Bio, and Freenome; she consults for Neumora. SKN reports compensation for consulting services with Radera Biosciences. All other authors declare that they have no competing interests.
Figures





References
-
- Meek J. Y., Noble L., Section on Breastfeeding, Policy Statement: Breastfeeding and the Use of Human Milk. Pediatrics 150 (2022). - PubMed
-
- Breastfeeding. https://www.who.int/health-topics/breastfeeding.
-
- Breastfeeding, UNICEF DATA (2021). https://data.unicef.org/topic/nutrition/breastfeeding/.
-
- CDC, Results: Breastfeeding rates, Centers for Disease Control and Prevention; (2023). https://www.cdc.gov/breastfeeding/data/nis_data/results.html.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
