This is a preprint.
Missense variants in TUBA4A cause myo-tubulinopathies
- PMID: 40666348
- PMCID: PMC12262762
- DOI: 10.1101/2025.06.26.25330266
Missense variants in TUBA4A cause myo-tubulinopathies
Abstract
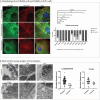
Tubulinopathies encompass a wide spectrum of disorders resulting from variants in genes encoding α- and β-tubulins, the key components of microtubules. While previous studies have linked de novo or dominantly inherited TUBA4A missense variants to neurodegenerative phenotypes, including amyotrophic lateral sclerosis, frontotemporal dementia, hereditary spastic ataxia, and more recently, an isolated report of congenital myopathy, the full phenotypic and genotypic spectrum of TUBA4A-related disorders remains incompletely characterised. In this multi-centre study, we identified 13 novel TUBA4A missense variants in 31 individuals from 19 unrelated families. Remarkably, affected individuals in 17 families presented with a primary axial myopathy without any identified CNS involvement or history of such disease. In the remaining two families, we observed probands with cerebellar ataxia and epilepsy accompanying proximal and axial muscle weakness, establishing the first documented association between TUBA4A variants and multisystem proteinopathy. Our cohort exhibited diverse genotypes and associated inheritance patterns: four families demonstrated autosomal dominant transmission through heterozygous variants in TUBA4A, three probands had homozygous TUBA4A variants, where the biallelic genotype was found to be associated with the disease, and the heterozygous carriers were asymptomatic; five probands carried de novo variants, and nine probands with heterozygous TUBA4A variants were classified as "isolated-sporadic cases" where parental samples were unavailable. Clinical phenotypes ranged from mild to severe myopathy, predominantly affecting the axial and paraspinal muscles. We observed a range of disease onset, from congenital to late adulthood. Creatine kinase levels were also variable, ranging from normal to highly elevated. Cardiac function remained preserved across the cohort. Muscle biopsies revealed a range of pathologies, including myofibre size variation, myofibre atrophy, nemaline bodies, core-like regions, internal nuclei, and endomysial fibrosis. Immunohistochemical staining showed evidence of proteinopathy, with autophagic features and TUBA4A accumulation in patient myofibres. Complementary in silico and in vitro investigations suggested that the identified TUBA4A substitutions cause significant protein abnormalities and may differentially impact microtubule dynamics. Our findings establish myo-tubulinopathies as distinct clinical entities, encompassing both primary myopathies and multisystem proteinopathies with muscle involvement. This study broadens the phenotypic and genotypic spectrum of TUBA4A-related disorders beyond autosomal dominant or de novo mechanisms and neurodegenerative presentations. These results underscore the importance of considering TUBA4A variants in the differential diagnosis of axial myopathies and multisystem proteinopathies, regardless of central nervous system (CNS) involvement.
Keywords: autophagy; genotype-phenotype correlation; protein aggregate myopathy; tubulin; tubulinopathy.
Conflict of interest statement
Competing interests The authors report no competing interests.
Figures



References
-
- Bahi-Buisson N, Maillard C. Tubulinopathies Overview. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, eds. GeneReviews((R)). University of Washington, Seattle - PubMed
-
Copyright © 1993–2024, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.; 1993.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
