T-cell repertoire correlates with cytokine imbalance in multiple sclerosis patients
- PMID: 40666507
- PMCID: PMC12259453
- DOI: 10.3389/fimmu.2025.1604452
T-cell repertoire correlates with cytokine imbalance in multiple sclerosis patients
Abstract
Indroduction: Multiple sclerosis (MS) is mediated by innate and adaptive immune response deviation involving immune cells and cytokines. Here, we investigated whether combined cytokine profiling and T-cell receptor (TCR) repertoire analysis can better display the complex landscape of MS-driving immune responses.
Methods: We used advanced computational methods to systematically cluster highly variable individual levels of 48 cytokines in cerebrospinal fluid (CSF) and blood of 24 MS patients compared to that of nine controls. Relevant TCR sequences were compared to 88 healthy controls. We correlated cytokines with predominant shared TCR sequences to identify immune response networks.
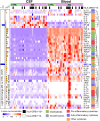
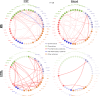
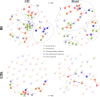
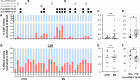
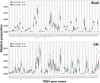
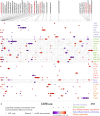
Results: MS patients had significantly elevated MIP-1α and IP-10 levels in CSF, and additional 36 blood cytokines variably but significantly elevated. We identified 77 predominantly pro-inflammatory cytokine correlations in MS-CSF. TCR sequencing revealed more productive rearrangements in CSF of MS and a significantly higher shared clone recovery rate in blood. We found significant associations involving 492 unique sequences and 34 cytokines in blood. Particularly, the less significant individual cytokine deviations were found to contribute to a general Th1-biased type I immune response correlating with clonal expansion of T cells directed against EBV, CMV, and other infectious agents.
Discussion: Correlation of significantly altered T-cell repertoire with cytokine deviations in MS despite individual patient data variability indicates that future diagnostic strategies may need to address immune response patterns rather than individual protein targets.
Keywords: T cell repertoire; bioinformatics; cytokine imbalance; multiple sclerosis; neuroscience.
Copyright © 2025 Weidner, Poupardin, Zrzavy, Laner-Plamberger, Gratz, Eichhorn, Weber, Rommer, Jungbauer and Strunk.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

