A novel post-mortem pathogen discovery program detects an outbreak of Echovirus E7: Uganda, 2022-2023
- PMID: 40666800
- PMCID: PMC12259650
- DOI: 10.3389/fmicb.2025.1557576
A novel post-mortem pathogen discovery program detects an outbreak of Echovirus E7: Uganda, 2022-2023
Abstract
Objectives: Utilizing post-mortem examination for routine monitoring of infectious diseases and pandemic preparedness is a common-sense, yet uncommon, public health measure. Here, we established a novel mortuary surveillance program in Uganda that leverages the unbiased nature of metagenomic next-generation sequencing (mNGS) to detect pathogens in recently deceased individuals.
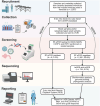
Methods: Between October 2022 and December 2023, specimens and patient metadata were collected from 2,607 individuals across five mortuary sites around Kampala. Specimens were pre-screened for hemorrhagic fever viruses by RT-qPCR and a subset (n = 134) of RT-qPCR negatives were sequenced by mNGS.
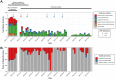
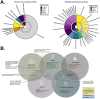
Results: A variety of DNA (herpes, parvovirus, bufavirus) and RNA (Saffold, Salivirus, HAV) viruses, vectored (Bartonella, Rickettsia) and nosocomial (Enterobacter, Klebsiella) bacterial infections, and potentially lethal respiratory pathogens (e.g., Cryptococcus neoformans, Corynebacterium diphtheria) were detected. A localized outbreak of Enterovirus B (EV-B), specifically a recombinant Echovirus E7, was observed in Kampala. An epidemiologic assessment indicated that most identified pathogens were acquired via direct and/or indirect contact (e.g., fecal-oral, fomites) and that other modes of transmission (e.g., food-borne, insect-vectored) played a less significant role.
Conclusion: Integration of mortuary surveillance, coupled with mNGS, into public health systems represents a powerful strategy for identifying unrecognized outbreaks and monitoring the (re-) emergence of infectious diseases.
Keywords: Uganda; echovirus; enterovirus; metagenomics; mortuary surveillance.
Copyright © 2025 Weiss, Bbosa, Orf, Berg, Ssemwanga, Kalungi, Balinandi, Mata, Bosa, Nabirye, Buule, Lutalo, Havron, Downing, Rodgers, Averhoff, Cloherty and Kaleebu.
Conflict of interest statement
SW, GO, MB, MM, AH, MR, FA, and GC are Abbott employees and shareholders. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Aceng J. R., Bosa H. K., Kamara N., Atwine D., Mwebesa H., Nyika H., et al. (2023). Continental concerted efforts to control the seventh outbreak of Ebola virus disease in Uganda: the first 90 days of the response. J. Public Health Afr. 14:2735. doi: 10.4081/jphia.2023.2735, PMID: - DOI - PMC - PubMed
-
- Ashraf S., Jerome H., Davis C., Shepherd J., Vattipally S., Filipe A., et al. (2023). Emerging viruses are an underestimated cause of undiagnosed febrile illness in Uganda. Int. J. Infect. Dis. 130:S17. doi: 10.1101/2023.04.27.23288465 - DOI
LinkOut - more resources
Full Text Sources

