This is a preprint.
Epigenetic Programming of Estrogen Receptor in Adipose Tissue by High Fat Diet Regulates Obesity-Induced Inflammation
- PMID: 40666843
- PMCID: PMC12262827
- DOI: 10.1101/2025.06.21.660886
Epigenetic Programming of Estrogen Receptor in Adipose Tissue by High Fat Diet Regulates Obesity-Induced Inflammation
Update in
-
Epigenetic programming of estrogen receptor in adipocytes by high-fat diet regulates obesity-induced inflammation.JCI Insight. 2025 Aug 26;10(19):e173423. doi: 10.1172/jci.insight.173423. eCollection 2025 Oct 8. JCI Insight. 2025. PMID: 40857119 Free PMC article.
Abstract
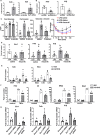
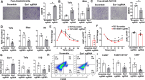
Adipose inflammation plays a key role in obesity-induced metabolic abnormalities. Epigenetic regulation, including DNA methylation, is a molecular link between environmental factors and complex diseases. Here we found that high fat diet (HFD) feeding induced a dynamic change of DNA methylome in mouse white adipose tissue (WAT) analyzed by reduced representative bisulfite sequencing. Interestingly, DNA methylation at the promoter of estrogen receptor α (Esr1) was significantly increased by HFD, concomitant with a down-regulation of Esr1 expression. HFD feeding in mice increased the expression of DNA methyltransferase 1 (Dnmt1) and Dnmt3a, and binding of DNMT1 and DNMT3a to Esr1 promoter in WAT. Mice with adipocyte-specific Dnmt1 deficiency displayed increased Esr1 expression, decreased adipose inflammation and improved insulin sensitivity upon HFD challenge; while mice with adipocyte-specific Dnmt3a deficiency showed a mild metabolic phenotype. Using a modified CRISPR/RNA-guided system to specifically target DNA methylation at the Esr1 promoter in WAT, we found that reducing DNA methylation at Esr1 promoter increased Esr1 expression, decreased adipose inflammation and improved insulin sensitivity in HFD-challenged mice. Our study demonstrates that DNA methylation at Esr1 promoter plays an important role in regulating adipose inflammation, which may contribute to obesity-induced insulin resistance.
Keywords: Adipose inflammation; DNA methylation; Estrogen receptor.
Conflict of interest statement
Conflicts of Interest: The authors have no conflict of interest to declare.
Figures




References
-
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–7. - PubMed
-
- Olefsky JM, and Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol.72:219–46. - PubMed
-
- Suzuki MM, and Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–76. - PubMed
-
- Backdahl L, Bushell A, and Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int J Biochem Cell Biol. 2009;41(1):176–84. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
