This is a preprint.
Linker Length and Composition within Disordered Binding Motifs modulates the Avidity and Reversibility of a Multivalent Protein Interaction Switch
- PMID: 40666912
- PMCID: PMC12262211
- DOI: 10.1101/2025.06.06.658374
Linker Length and Composition within Disordered Binding Motifs modulates the Avidity and Reversibility of a Multivalent Protein Interaction Switch
Abstract
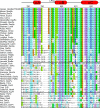
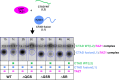
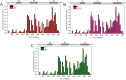
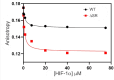
Intrinsically disordered proteins that mediate the cellular transcriptional response to hypoxia play important roles in turning on and turning off oxygen stress genes. In particular, the feedback inhibitor CITED2 operates a unidirectional switch that efficiently terminates the hypoxic response by displacing the C-terminal activation domain of the hypoxia-inducible factor HIF-1α from its complex with the TAZ1 domain of the transcriptional coactivators CBP and p300. Unidirectionality of the switch arises from subtle allosteric conformational changes in TAZ1 and from differences in the strength of thermodynamic coupling between the TAZ1-binding motifs in the multivalent HIF-1α and CITED2 activation domains. To investigate the role of binding cooperativity, we mutated a linker sequence in the HIF-1α activation domain to enhance or reduce the thermodynamic coupling between its TAZ1-binding motifs. An efficient native-gel assay shows that certain linker mutations enhance the affinity of HIF-1α for TAZ1, and fluorescence anisotropy competition and NMR measurements show that these mutants can compete with CITED2 for TAZ1 more effectively than wild-type HIF-1α. The wide range of mutants, which include insertion, deletion, replacement and scrambling of residues in the linker, provide insights into the molecular basis for the exquisite tuning of the hypoxic switch: the TAZ1 affinity and consequent CITED2 competition enhancement depends both on the flexibility of the linker sequence (particularly the presence of glycine residues) and the unfavorable electrostatic interactions of a highly conserved arginine side chain in the center of the linker with an electropositive surface of TAZ1.
Figures




Similar articles
-
Hypersensitive termination of the hypoxic response by a disordered protein switch.Nature. 2017 Mar 16;543(7645):447-451. doi: 10.1038/nature21705. Epub 2017 Mar 8. Nature. 2017. PMID: 28273070 Free PMC article.
-
Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.J Biol Chem. 2004 Jan 23;279(4):3042-9. doi: 10.1074/jbc.M310348200. Epub 2003 Oct 31. J Biol Chem. 2004. PMID: 14594809
-
Multivalency enables unidirectional switch-like competition between intrinsically disordered proteins.Proc Natl Acad Sci U S A. 2022 Jan 18;119(3):e2117338119. doi: 10.1073/pnas.2117338119. Proc Natl Acad Sci U S A. 2022. PMID: 35012986 Free PMC article.
-
Hypoxia-inducible factor 1alpha and vascular endothelial growth factor in Glioblastoma Multiforme: a systematic review going beyond pathologic implications.Oncol Res. 2024 Jul 17;32(8):1239-1256. doi: 10.32604/or.2024.052130. eCollection 2024. Oncol Res. 2024. PMID: 39055895 Free PMC article.
-
Effect of hypoxia-inducible factor 1 on vascular endothelial growth factor expression in exercised human skeletal muscle: a systematic review and meta-analysis.Am J Physiol Cell Physiol. 2025 Jul 1;329(1):C272-C282. doi: 10.1152/ajpcell.00297.2025. Epub 2025 Jun 16. Am J Physiol Cell Physiol. 2025. PMID: 40522862 Review.
References
-
- Sipko E. L., Chappell G. F., and Berlow R. B. (2024) Multivalency emerges as a common feature of intrinsically disordered protein interactions, Curr. Opin. Struct. Biol. 84, 102742. - PubMed
-
- Weng J., and Wang W. (2020) Dynamic multivalent interactions of intrinsically disordered proteins, Curr. Opin. Struct. Biol. 62, 9–13. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
