This is a preprint.
Inhibition of nonsense-mediated decay in TDP-43 deficient neurons reveals novel cryptic exons
- PMID: 40667039
- PMCID: PMC12262666
- DOI: 10.1101/2025.06.28.661837
Inhibition of nonsense-mediated decay in TDP-43 deficient neurons reveals novel cryptic exons
Abstract
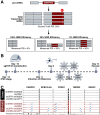
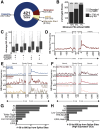
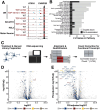
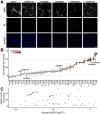
TAR DNA-binding protein 43 kDa (TDP-43) is an essential splicing repressor whose loss of function underlies the pathophysiology of amyotrophic lateral sclerosis and frontotemporal dementia (ALS-FTD). Nuclear clearance of TDP-43 disrupts its function and leads to the inclusion of aberrant cryptic exons. These cryptic exons frequently introduce premature termination codons resulting in the degradation of affected transcripts through nonsense-mediated mRNA decay (NMD). Conventional RNA sequencing approaches thus may fail to detect cryptic exons that are efficiently degraded by NMD, precluding identification of potential therapeutic targets. We generated a comprehensive set of neuronal targets of TDP-43 in human iPSC-derived i3Neurons (i3N) by combining TDP-43 knockdown with inhibition of multiple factors essential for NMD, revealing novel cryptic targets. We then restored expression of selected NMD targets in TDP-43 deficient i3Ns and determined which genes improved neuronal viability. Our findings highlight the role of NMD in masking cryptic splicing events and identify novel potential therapeutic targets for TDP-43-related neurodegenerative disorders.
Figures

Similar articles
-
Long-read RNA sequencing unveils a novel cryptic exon in MNAT1 along with its full-length transcript structure in TDP-43 proteinopathy.Commun Biol. 2025 Jul 16;8(1):1056. doi: 10.1038/s42003-025-08463-4. Commun Biol. 2025. PMID: 40670663 Free PMC article.
-
Cryptic splicing of stathmin-2 and UNC13A mRNAs is a pathological hallmark of TDP-43-associated Alzheimer's disease.Acta Neuropathol. 2024 Jan 4;147(1):9. doi: 10.1007/s00401-023-02655-0. Acta Neuropathol. 2024. PMID: 38175301 Free PMC article.
-
Large-scale RNA-Seq mining reveals ciclopirox olamine induces TDP-43 cryptic exons.Nat Commun. 2025 Jul 25;16(1):6878. doi: 10.1038/s41467-025-62004-5. Nat Commun. 2025. PMID: 40715064 Free PMC article.
-
TARDBP-Related Amyotrophic Lateral Sclerosis-Frontotemporal Dementia.2009 Apr 23 [updated 2023 Jan 5]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2009 Apr 23 [updated 2023 Jan 5]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301761 Free Books & Documents. Review.
-
Decoding TDP-43: the molecular chameleon of neurodegenerative diseases.Acta Neuropathol Commun. 2024 Dec 31;12(1):205. doi: 10.1186/s40478-024-01914-9. Acta Neuropathol Commun. 2024. PMID: 39736783 Free PMC article. Review.
References
-
- Neumann M. et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 314, 130–133 (2006). - PubMed
-
- Arai T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
