This is a preprint.
Phosphoglycerate Kinase Can Adopt a Topologically Misfolded Form that is More Stable than its Native State
- PMID: 40667093
- PMCID: PMC12262362
- DOI: 10.1101/2025.06.24.661412
Phosphoglycerate Kinase Can Adopt a Topologically Misfolded Form that is More Stable than its Native State
Abstract
The native states of globular proteins are typically viewed as being the most stable conformations on their respective proteins' soluble free energy landscapes. This view, known as the Thermodynamic Hypothesis, explains why many proteins can reversibly refold after being denatured. Here we report an intriguing counterexample to this paradigm. When E. coli phosphoglycerate kinase (PGK) is stimulated to refold upon dilution from denaturant, instead of returning to its native state, it populates an unusual misfolded form that is monomeric and native-like, but which is even more kinetically stable than its native form, as based on its resistance to thermal and detergent-induced denaturation. Moreover, this misfolded form cannot self-correct, even for days. We show that the key structural feature of this misfolded form of PGK is topological in nature by demonstrating that kinetically stable misfolded forms do not form any longer if PGK is circularized, which prevents its termini from threading through other portions of the protein. Our findings demonstrate that a misfolded protein need not aggregate or form an amyloid to become stabilized with respect to the native state, and call attention to topologically-misfolded proteins as a potential Achilles heel to the cellular proteostasis network.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

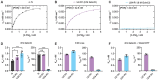



Similar articles
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
"In a State of Flow": A Qualitative Examination of Autistic Adults' Phenomenological Experiences of Task Immersion.Autism Adulthood. 2024 Sep 16;6(3):362-373. doi: 10.1089/aut.2023.0032. eCollection 2024 Sep. Autism Adulthood. 2024. PMID: 39371355
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Anfinsen C. B., Principles that Govern the Folding of Protein Chains. Sci. New Ser. 181, 223–230 (1973). - PubMed
-
- Dill K. A., Chan H. S., From Levinthal to pathways to funnels. Nat. Struct. Mol. Biol. 4, 10–19 (1997). - PubMed
-
- Karplus M., Behind the folding funnel diagram. Nat. Chem. Biol. 7, 401–404 (2011). - PubMed
-
- Hartl F. U., Hayer-Hartl M., Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 (2009). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
