This is a preprint.
Divide and Cluster: The DIVINE Framework for Deterministic Top-Down Analysis of Molecular Dynamics Trajectories
- PMID: 40667114
- PMCID: PMC12262464
- DOI: 10.1101/2025.06.20.660828
Divide and Cluster: The DIVINE Framework for Deterministic Top-Down Analysis of Molecular Dynamics Trajectories
Abstract
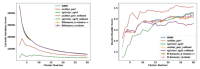
We present DIVIsive N -ary Ensembles (DIVINE), a deterministic, top-down clustering framework designed for molecular dynamics (MD) trajectories. DIVINE constructs a complete clustering hierarchy by recursively splitting clusters based on n -ary similarity principles, avoiding the need for O(N 2 ) pairwise distance matrices. It supports multiple cluster selection criteria, including a weighted variance metric, and deterministic anchor initialization strategies such as NANI (N-ary Natural Initiation), ensuring reproducible and well-balanced partitions. Testing DIVINE up to a 305 μs folding trajectory of the villin headpiece (HP35) revealed that it matched or exceeded the clustering quality of bisecting k -means while reducing runtime and eliminating stochastic variability. Its single-pass design enables efficient exploration of clustering resolutions without repeated executions. By combining scalability, interpretability, and determinism, DIVINE offers a robust and practical alternative to conventional MD clustering methods. DIVINE is publicly available as part of the MDANCE package: https://github.com/mqcomplab/MDANCE .
Conflict of interest statement
Conflict of Interest: The authors declare no competing financial interests.
Figures


Similar articles
-
Scaling k-Means for Multi-Million Frames: A Stratified NANI Approach for Large-Scale MD Simulations.bioRxiv [Preprint]. 2025 Jun 18:2025.06.15.659780. doi: 10.1101/2025.06.15.659780. bioRxiv. 2025. PMID: 40666979 Free PMC article. Preprint.
-
Hierarchical Extended Linkage Method (HELM)'s Deep Dive into Hybrid Clustering Strategies.bioRxiv [Preprint]. 2025 Mar 10:2025.03.05.641742. doi: 10.1101/2025.03.05.641742. bioRxiv. 2025. Update in: J Chem Inf Model. 2025 Jun 23;65(12):6209-6220. doi: 10.1021/acs.jcim.5c00539. PMID: 40161705 Free PMC article. Updated. Preprint.
-
Hierarchical Extended Linkage Method (HELM)'s Deep Dive into Hybrid Clustering Strategies.J Chem Inf Model. 2025 Jun 23;65(12):6209-6220. doi: 10.1021/acs.jcim.5c00539. Epub 2025 Jun 2. J Chem Inf Model. 2025. PMID: 40452401
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Workplace interventions for reducing sitting at work.Cochrane Database Syst Rev. 2016 Mar 17;3(3):CD010912. doi: 10.1002/14651858.CD010912.pub3. Cochrane Database Syst Rev. 2016. Update in: Cochrane Database Syst Rev. 2018 Jun 20;6:CD010912. doi: 10.1002/14651858.CD010912.pub4. PMID: 26984326 Free PMC article. Updated.
References
-
- Arnittali M.; Rissanou A. N.; Harmandaris V. Structure Of Biomolecules Through Molecular Dynamics Simulations. Procedia Computer Science 2019, 156, 69–78. 10.1016/j.procs.2019.08.181. - DOI
-
- Shaw D. E.; Deneroff M. M.; Dror R. O.; Kuskin J. S.; Larson R. H.; Salmon J. K.; Young C.; Batson B.; Bowers K. J.; Chao J. C.; Eastwood M. P.; Gagliardo J.; Grossman J. P.; Ho C. R.; Ierardi D. J.; Kolossváry I.; Klepeis J. L.; Layman T.; McLeavey C.; Moraes M. A.; Mueller R.; Priest E. C.; Shan Y.; Spengler J.; Theobald M.; Towles B.; Wang S. C. Anton, a Special-Purpose Machine for Molecular Dynamics Simulation. Commun. ACM 2008, 51 (7), 91–97. 10.1145/1364782.1364802. - DOI
Publication types
LinkOut - more resources
Full Text Sources
