This is a preprint.
Bidirectional regulation of glycoprotein nonmetastatic melanoma protein B by β-glucocerebrosidase deficiency in GBA1 isogenic dopaminergic neurons from a patient with Gaucher disease and parkinsonism
- PMID: 40667145
- PMCID: PMC12262256
- DOI: 10.1101/2025.06.23.661126
Bidirectional regulation of glycoprotein nonmetastatic melanoma protein B by β-glucocerebrosidase deficiency in GBA1 isogenic dopaminergic neurons from a patient with Gaucher disease and parkinsonism
Abstract
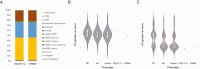
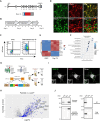
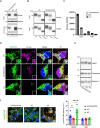
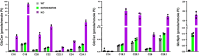
Variants in GBA1 are common genetic risk factors for several synucleinopathies. The increased risk has been attributed to the toxic effects of misfolded glucocerebrosidase (GCase) (gain-of-function), and the accumulation of lipid substrates due to reduced enzyme activity (loss-of-function). To delineate GBA1 pathogenicity, an iPSC line was generated from a patient with both type 1 Gaucher disease (GBA1: N370S/N370S; p.N409S/p.N409S) and Parkinson disease (PD). From this line, we created a reverted wild-type (WT) line and a GBA1 knockout (KO) line to eliminate misfolded GCase and intensify lipid accumulation. N370S/N370S and KO dopaminergic neurons (DANs) exhibited decreasing GCase levels and progressive accumulation of lipid substrates compared to WT DANs. Notably, the expression of GPNMB, whose levels correlate with PD risk, was upregulated by the mild lipid accumulation in N370S/N370S DANs but disrupted in KO DANs. These findings refine the loss-of-function mechanism by associating PD risk levels of GPNMB with lipid levels specific to GBA1 risk variants.
Keywords: GBA1; GCase; GPNMB; Gaucher disease; Parkinson disease; iPSC.
Conflict of interest statement
Relevant conflicts of interest: Z.L.’s participation in this project was part of a competitive contract awarded to DataTecnica International LLC by the NIH to support open science research.
Figures


Similar articles
-
Comparative study of enriched dopaminergic neurons from siblings with Gaucher disease discordant for parkinsonism.bioRxiv [Preprint]. 2024 Feb 28:2024.02.25.581985. doi: 10.1101/2024.02.25.581985. bioRxiv. 2024. Update in: Mov Disord. 2025 Jun 26. doi: 10.1002/mds.30273. PMID: 38529501 Free PMC article. Updated. Preprint.
-
Novel beta-glucocerebrosidase chaperone compounds identified from cell-based screening reduce pathologically accumulated glucosylsphingosine in iPS-derived neuronal cells.SLAS Discov. 2023 Oct;28(7):344-349. doi: 10.1016/j.slasd.2023.06.002. Epub 2023 Jun 25. SLAS Discov. 2023. PMID: 37369311
-
Evaluation of Induced Pluripotent Stem Cell-Derived Dopaminergic Neurons from Siblings with Gaucher Disease Discordant for Parkinsonism.Mov Disord. 2025 Jun 26. doi: 10.1002/mds.30273. Online ahead of print. Mov Disord. 2025. PMID: 40568761
-
Classification and Genotype-Phenotype Relationships of GBA1 Variants: MDSGene Systematic Review.Mov Disord. 2025 Apr;40(4):605-618. doi: 10.1002/mds.30141. Epub 2025 Feb 10. Mov Disord. 2025. PMID: 39927608 Free PMC article.
-
Frequency of Hereditary and GBA1-Related Parkinsonism in Latin America: A Systematic Review and Meta-Analysis.Mov Disord. 2024 Jan;39(1):6-16. doi: 10.1002/mds.29614. Epub 2023 Nov 3. Mov Disord. 2024. PMID: 37921246
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
