This is a preprint.
Harnessing fusion of genome-edited human stem cells to rapidly screen for novel protein functions in vivo
- PMID: 40667162
- PMCID: PMC12262239
- DOI: 10.1101/2025.06.25.661608
Harnessing fusion of genome-edited human stem cells to rapidly screen for novel protein functions in vivo
Update in
-
Harnessing fusion of genome-edited human stem cells to rapidly screen for novel protein functions in vivo.Mol Biol Cell. 2025 Nov 1;36(11):ar141. doi: 10.1091/mbc.E25-06-0301. Epub 2025 Sep 24. Mol Biol Cell. 2025. PMID: 40991409
Abstract
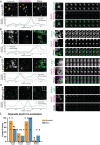
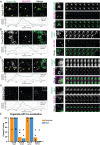
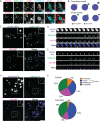
Genome editing has enabled the integration of fluorescent protein coding sequences into genomes, resulting in expression of in-frame fusion proteins under the control of their natural gene regulatory sequences. While this technique overcomes the well-documented artifacts associated with gene overexpression, editing genomes of metazoan cells incurs a significant time cost compared to simpler organisms, such as yeast. Editing two or more genes to express multiple fluorescent fusion proteins in a single cell line has proven to be a powerful strategy for uncovering spatio-dynamic, and therefore functional, relationships among different proteins, but it can take many months to edit each gene within the same cell line. Here, by utilizing cell fusions, we quickly generated cells expressing pairwise permutations of fluorescent fusion proteins in genome-edited human cells to reveal previously undetected protein-organelle interactions. We fused human induced pluripotent stem cells (hiPSCs) that express in-frame fusions of clathrin-mediated endocytosis (CME) and actin cytoskeleton proteins with hiPSCs that express fluorescently tagged organelle markers, uncovering novel interactions between CME proteins, branched actin filament networks, and lysosomes.
Conflict of interest statement
Competing Interests The authors declare no competing interest.
Figures


References
-
- Brodsky F. M. (2016). Clathrin and Clathrin-Dependent Endocytosis. In Encyclopedia of Cell Biology (pp. 384–393). Elsevier. 10.1016/B978-0-12-394447-4.20038-2 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
