This is a preprint.
Spatially-distinct programming of macrophage diversity within the granulomas of Mycobacterium tuberculosis infected nonhuman primates
- PMID: 40667206
- PMCID: PMC12262409
- DOI: 10.1101/2025.06.12.659348
Spatially-distinct programming of macrophage diversity within the granulomas of Mycobacterium tuberculosis infected nonhuman primates
Abstract
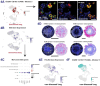
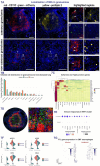
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is defined by granulomas-immune aggregates that either contain or support bacterial replication. Macrophages, fundamental components of these lesions, are crucial to TB pathogenesis, yet their phenotypic and functional diversity is incompletely understood. Here, we used single-cell RNA sequencing and immunofluorescence to profile macrophages in lung tissue and granulomas from a nonhuman primate model of early TB. We identified distinct subsets, including embryonic-origin tissue-resident alveolar macrophages and monocyte-derived alveolar and interstitial macrophages, with distinct spatial localization in granulomas. Tissue-resident alveolar macrophages and a subset undergoing epithelial-to-mesenchymal transition accounted for the highest frequency of Mtb-infected cells. Infected cells exhibited differential expression of immune- and migration-associated genes compared to uninfected counterparts, suggesting Mtb either induces or exploits these pathways as a survival strategy. These findings highlight macrophage heterogeneity as a major driver of differential susceptibility to Mtb and provide insights relevant to future immunomodulatory strategies.
Conflict of interest statement
Conflict of Interest: The authors declare no conflicts of interest in this work.
Figures





References
-
- WHO. 2023. [cited 2024 April 22]; Available from: https://www.who.int/news-room/fact-sheets/detail/tuberculosis.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
