This is a preprint.
The soluble HIV-1 Vpu protein interacts with calmodulin in a Ca2+-dependent manner
- PMID: 40667306
- PMCID: PMC12262824
- DOI: 10.1101/2025.06.12.658902
The soluble HIV-1 Vpu protein interacts with calmodulin in a Ca2+-dependent manner
Update in
-
The soluble state of the HIV-1 Vpu protein forms a complex with Ca2+-calmodulin.Protein Sci. 2026 Feb;35(2):e70487. doi: 10.1002/pro.70487. Protein Sci. 2026. PMID: 41575075
Abstract
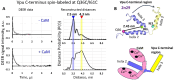
The HIV-1-encoded membrane protein Vpu plays key roles in virus lifecycle. Our lab recently revealed a soluble form of Vpu, and we strived to determine its possible physiological function. Here, we provide solid experimental proof that soluble Vpu interacts with Ca2+-bound calmodulin (Ca2+-CaM). A putative CaM-binding motif in Vpu was predicted, but there was no experimental evidence of the Vpu-CaM association. We applied double electron electron-resonance (DEER) and protein spin labeling to detect the soluble Vpu-CaM complex. We found that soluble full-length and truncated C-terminal region of Vpu directly interact with Ca2+-CaM. DEER results from the spin-labeled double cysteine mutant S39C/A103C of CaM showed that upon association with Vpu Ca2+-CaM adopts a more closed conformation compared to those in the absence of Vpu. This restructuring is in agreement with previously observed Ca2+-CaM association with cellular and other HIV-1 proteins. Our results indicate that soluble Vpu and CaM form an equimolar complex. DEER results from doubly spin-labeled at residues Q36C/I61C in Vpu C-terminal region suggest that Vpu's helices 2 and 3 move away from each other to facilitated CaM binding. These observations tell that under physiological conditions the soluble Vpu-CaM complex may provide Vpu with a trafficking pathway to membrane destination.
Keywords: DEER spectroscopy to study protein-protein complexes; protein conformational changes in protein-protein interaction; soluble HIV-1 Vpu protein; soluble Vpu-calmodulin interaction.
Figures



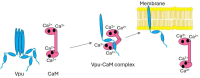
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
