This is a preprint.
Extracellular Vesicle-Mediated Delivery of Genetic Material for Transformation and CRISPR/Cas9-based Gene Editing in Pneumocystis murina
- PMID: 40667333
- PMCID: PMC12262392
- DOI: 10.1101/2025.06.17.660080
Extracellular Vesicle-Mediated Delivery of Genetic Material for Transformation and CRISPR/Cas9-based Gene Editing in Pneumocystis murina
Abstract
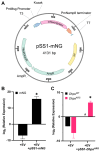
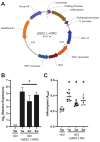
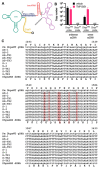
Pneumocystis species are obligate fungal pathogens that cause severe pneumonia, particularly in immunocompromised individuals. The absence of robust genetic manipulation tools has impeded our mechanistic understanding of Pneumocystis biology and the development of novel therapeutic strategies. Herein, we describe a novel method for the stable transformation and CRISPR/Cas9-mediated genetic editing of Pneumocystis murina utilizing extracellular vesicles (EVs) as a delivery vehicle. Building upon our prior investigations demonstrating EV-mediated delivery of exogenous material to Pneumocystis, we engineered mouse lung EVs to deliver plasmid DNA encoding reporter genes and CRISPR/Cas9 components. Our initial findings demonstrated successful in vitro transformation and subsequent expression of mNeonGreen and DhpsARS in P. murina organisms. Subsequently, we established stable in vivo expression of mNeonGreen in mice infected with transformed P. murina for a duration of up to 5 weeks. Furthermore, we designed and validated a CRISPR/Cas9 system targeting the P. murina Dhps gene, confirming its in vitro cleavage efficiency. Ultimately, we achieved successful in vivo CRISPR/Cas9-mediated homologous recombination, precisely introducing a DhpsARS mutation into the P. murina genome, which was confirmed by Sanger sequencing across all tested animals. Here, we establish a foundational methodology for genetic manipulation in Pneumocystis, thereby opening avenues for functional genomics, drug target validation, and the generation of genetically modified strains for advanced research and potential therapeutic applications.
Figures


Similar articles
-
124I-labelled BMSC-Derived Extracellular Vesicles Deliver CRISPR/Cas9 Ribonucleoproteins With a GFP-Reporter System to Inhibit Osteosarcoma Proliferation and Metastasis.J Extracell Vesicles. 2025 Jul;14(7):e70130. doi: 10.1002/jev2.70130. J Extracell Vesicles. 2025. PMID: 40673793 Free PMC article.
-
Modulating binding affinity of aptamer-based loading constructs enhances extracellular vesicle-mediated CRISPR/Cas9 delivery.J Control Release. 2025 Aug 10;384:113853. doi: 10.1016/j.jconrel.2025.113853. Epub 2025 May 18. J Control Release. 2025. PMID: 40393529
-
Harnessing an anti-CRISPR protein for powering CRISPR/Cas9-mediated genome editing in undomesticated Bacillus strains.Microb Cell Fact. 2025 Jun 23;24(1):143. doi: 10.1186/s12934-025-02776-z. Microb Cell Fact. 2025. PMID: 40551141 Free PMC article.
-
Transferable approaches to CRISPR-Cas9 induced genome editing in non-model insects: a brief guide.Front Zool. 2025 Jul 7;22(1):13. doi: 10.1186/s12983-025-00566-2. Front Zool. 2025. PMID: 40624545 Free PMC article. Review.
-
CRISPR/Cas9-mediated genome editing in Ganoderma lucidum: recent advances and biotechnological opportunities.World J Microbiol Biotechnol. 2025 Jun 25;41(7):223. doi: 10.1007/s11274-025-04458-9. World J Microbiol Biotechnol. 2025. PMID: 40560492 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
