Exploration of fecal microbiota in newly diagnosed patients with inflammatory bowel disease using shotgun metagenomics
- PMID: 40667420
- PMCID: PMC12259665
- DOI: 10.3389/fcimb.2025.1595884
Exploration of fecal microbiota in newly diagnosed patients with inflammatory bowel disease using shotgun metagenomics
Abstract
Introduction: Dysbiosis is a key mechanism in inflammatory bowel disease (IBD) pathophysiology. Previous microbiota studies in IBD generally have involved patients treated with immunosuppressive agents, which can affect the results. We aimed to elucidate the fecal microbiota composition in newly diagnosed treatment-naïve IBD patients.
Methods: Microbiota from stool samples were investigated using shotgun metagenomics sequencing and subsequent bioinformatics analysis.
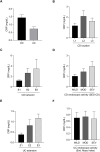
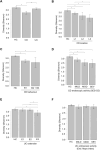
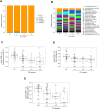
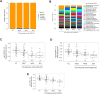
Results: A total of 103 patients with Crohn's disease (CD), 144 with ulcerative colitis (UC), and 49 healthy controls (HC) were included. CD patients had significantly lower species-level diversity than those with UC and HC. CD subgroups with Ileocolonic location and stricturing behavior showed reduced diversity compared to HC. A negative correlation was observed between endoscopic severity and microbial diversity in CD patients. UC patients had similar microbial diversity to HC, which was unaffected by disease activity. Taxonomic abundance analysis revealed a tendency towards a higher relative abundance of Escherichia coli and a lower relative abundance of Faecalibacterium prausnitzii in IBD patients compared to HC. However, the most significant differences in these patients compared to HC were observed in less abundant species, such as Toxoplasma gondii, Gemella morbillorum, and several species of the Adlercreutzia genera. Functional analysis in these patients highlighted changes in carbohydrate and nucleotide pathways.
Discussion: Our data suggest that newly diagnosed CD patients show significant microbiota composition disparities compared to UC patients and HC. Microbiota differences in these patients are linked to dysbiosis, characterized by a reduction in beneficial genera such as Gemella and Adlercreutzia, and a rise in pathogenic species.
Keywords: Crohn’s disease; inflammatory bowel disease; metagenomics; microbiota; shotgun; ulcerative colitis.
Copyright © 2025 Orejudo, Gómez, Riestra, Rivero, Gutiérrez, Rodríguez-Lago, Fernández-Salazar, Ceballos, Benítez, Aguas, Bastón-Rey, Bermejo, Casanova, Lorente-Poyatos, Ber, Ginard, Esteve, de Francisco, García, Francés, Rodríguez, Alcaide Suárez, Guerra del Río, Soto, Nos, Barreiro-de Acosta, Guerra, Hervías Cruz, Domínguez Cajal, Royo, Aceituno, Aldars-García, Garre, Ramírez, Soleto, Schuppe-Koistinen, Engstrand, Baldán-Martín, Sánchez-Cabo, Gisbert and Chaparro.
Conflict of interest statement
MC has served as speaker, consultant or research or education funding from MSD, Abbvie, Hospira, Pfizer, Takeda, Janssen, Ferring, Shire Pharmaceuticals, Dr. Falk Pharma, Tillotts Pharma, Biogen, Gilead and Lilly. JG has served as speaker, consultant, and advisory member for or has received research funding from MSD, Abbvie, Pfizer, Kern Pharma, Biogen, Mylan, Takeda, Janssen, Roche, Sandoz, Celgene/Bristol Myers, Gilead/Galapagos, Lilly, Ferring, Faes Farma, Shire Pharmaceuticals, Dr. Falk Pharma, Tillotts Pharma, Chiesi, Casen Fleet, Gebro Pharma, Otsuka Pharmaceutical, Norgine and Vifor Pharma. LF-S has served as advisory member for Ferring and has received research funding from Ferring, Janssen and Takeda. FB has been a speaker, consultant and advisory member or has received research funding from Abbvie, Takeda, Janssen, Pfizer, MSD, Biogen, Amgen, Galapagos, Ferring, Faes Farma, Tillotts Pharma, Chiesi, Vifor Pharma, Lilly. MB-A has served as a speaker, consultant and advisory member for or has received research funding from MSD, AbbVie, Janssen, Kern Pharma, Takeda, Galapagos-Alpha Sigma, Pfizer, Sandoz, Fresenius, Avibax, Lilly, Ferring, Faes Farma, Dr. Falk Pharma, Chiesi, Adacyte and TillotsPharma. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Andrews S. (2010). FastQC - A quality control tool for high throughput sequence data (Babraham Institute, Babraham, Cambridgeshire, United Kingdom: Babraham Bioinformatics; ). Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed May 11, 2022).
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

