A data-mining analysis of host solute carrier family proteins in SARS-CoV-2 infection with reference to brain endothelial cells and the blood-brain barrier in COVID-19
- PMID: 40667466
- PMCID: PMC12261995
- DOI: 10.3389/fneur.2025.1563040
A data-mining analysis of host solute carrier family proteins in SARS-CoV-2 infection with reference to brain endothelial cells and the blood-brain barrier in COVID-19
Abstract
Background: The brain vasculature is a key player in neurological manifestations of COVID-19. Infection of brain endothelial cells with SARS-CoV-2 along with circulating cytokines may cause dysfunction of the blood-brain barrier (BBB). Solute carrier transporters (SLCs) in brain endothelial cells regulate substrate transport across the BBB. Here, it was hypothesized that transport functions of SLCs will be impaired by interactions with viral proteins, and subsequently, data-mining studies were performed.
Methods: Virus-host protein-protein interaction data for SARS-CoV-2 infection were retrieved from the BioGRID database, filtered for SLCs, and then annotated for relevant expression in brain endothelial cells using a mouse brain transcriptomics database. Host SLCs expressed in brain endothelial cells were further explored using publicly available databases and information in the literature. Functional Annotation Clustering was performed using DAVID, and Enrichr served for pathway analysis. Substrates were retrieved from NCBI Gene. Links to monogenic disorders were retrieved from Online Mendelian Inheritance in Man™ and screened for disorders of the nervous system. Interactome data for viral proteins of SARS-CoV-2 were retrieved from BioGRID. Reports for host SLCs in viral receptor functions, viral entry mechanisms, and other major roles in the viral cycle were explored in databases (VThunter) and literature. ATP-binding cassette transporters (ABCs) were studied in parallel.
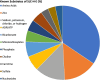
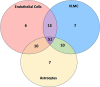
Results: N = 80 host SLCs showed relevant expression in brain endothelial cells whereby amino acid transporter stood out. N = 24/80 host SLCs were linked to monogenic disorders of the nervous system. N = 9/29 SARS-CoV-2 viral proteins had strong links to SLCs and key functions in viral infection (e.g., interferon response). SLCs serving as viral receptors and with closely associated functions were significantly enriched among all known listed viral receptors (chi-square test, p = 0.001). Literature searches for host SLCs revealed involvement of a subset of SLCs in infection mechanisms for SARS-CoV-2 and more broadly for other viruses. N = 17 host ABCs were found in brain endothelial cells where they may serve as efflux transporters.
Discussion: This hypothesis-generating work proposes a set of N = 80 host SLCs expressed in endothelial cells as contributors to BBB impairment after SARS-CoV-2 infection. Theoretically, persistent dysfunction of SLCs at the BBB, in particular insufficient transport of amino acids, could be one of many reasons for cognitive changes in long-COVID. Functions of SLCs in viral entry and associated roles deserve close attention.
Keywords: COVID-19; SARS-CoV-2; amino acid transport; blood-brain barrier; brain endothelial cells; protein-protein interactions; solute carrier proteins; virus-host interactions.
Copyright © 2025 Fradkin and Schmidt-Kastner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
References
LinkOut - more resources
Full Text Sources
Miscellaneous

