Influenza A virus induces PI4P production at the endoplasmic reticulum in an ATG16L1-dependent manner to promote the egress of viral ribonucleoproteins
- PMID: 40668844
- PMCID: PMC12286409
- DOI: 10.1371/journal.pbio.3002958
Influenza A virus induces PI4P production at the endoplasmic reticulum in an ATG16L1-dependent manner to promote the egress of viral ribonucleoproteins
Abstract
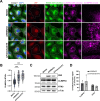
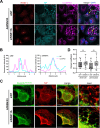
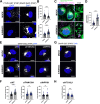
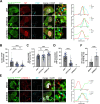
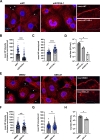
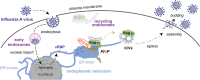
The genomic RNAs of influenza A viruses (IAVs) are replicated in the nucleus of infected cells in the form of viral ribonucleoproteins (vRNPs) before being exported to the cytoplasm. The small GTPase RAB11A is involved in the transport of vRNPs to the sites of viral assembly at the plasma membrane, but the molecular mechanisms involved remain largely unknown. Here we show that IAV infection remodels the architecture of the endoplasmic reticulum (ER) sheets, where vRNPs tend to accumulate in the absence of RAB11A. To decipher the interplay between RAB11A, vRNPs, and the ER, we investigated viral-induced perturbations of RAB11A proximity interactome. To this end, we generated cells stably expressing a TurboID-RAB11A fusion protein and performed biotin-based proximity labeling upon viral infection. We found that cellular regulators of phophatidylinositol-4-phosphate (PI4P) homeostasis, including the autophagic and stress response protein ATG16L1, are significantly enriched at the vicinity of RAB11A in infected cells. Infection induces an increase in cellular PI4P levels in an ATG16L1-dependent manner, while ATG16L1 relocalizes to ER membranes upon infection. Depletion of ATG16L1 decreases the co-distribution of vRNPs with PI4P punctae on ER membranes, and reduces the accumulation of vRNPs at the plasma membrane as well as the production of IAV infectious particles. Our data extend to IAVs the notion that viruses can modulate the metabolism and localization of phosphoinositides to control host membrane dynamics and point to the ER as an essential platform for vRNP transport. They provide evidence for a pivotal role of ATG16L1 in regulating the identity of endomembranes and coordinating RAB11A and PI4P-enriched membranes to ensure delivery of vRNPs to the plasma membrane.
Copyright: © 2025 Alemany et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

