Spatio-genetically coordinated TPR domain-containing proteins modulate c-di-GMP signaling in Vibrio vulnificus
- PMID: 40668869
- PMCID: PMC12282931
- DOI: 10.1371/journal.ppat.1013353
Spatio-genetically coordinated TPR domain-containing proteins modulate c-di-GMP signaling in Vibrio vulnificus
Abstract
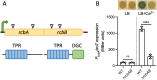
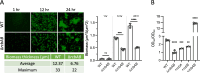
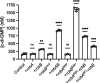
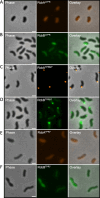
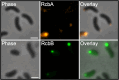
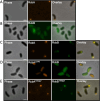
Vibrio species, which include several pathogens, are autochthonous to estuarine and warm coastal marine environments, where biofilm formation bolsters their ecological persistence and transmission. Here, we identify a bicistronic operon, rcbAB, whose products synergistically inhibit motility and promote biofilm maturation post-attachment by modulating intracellular c-di-GMP levels in the human and animal pathogen V. vulnificus. RcbA contains an N-terminal tetratricopeptide repeat (TPR) domain and a structured C-terminal region of unknown function, while RcbB possesses an N-terminal TPR domain and a C-terminal GGDEF domain characteristic of diguanylate cyclases. The TPR domain of RcbB represses its diguanylate cyclase activity, while RcbA's TPR domain and C-terminal region co-operatively de-repress it. Localization of both proteins to the flagellar pole is TPR-dependent but not co-dependent, although RcbA anchors RcbB to the pole in the absence of polar landmarks such as HubP and flagella. The conservation of rcbAB across diverse bacterial taxa substantiates its fundamental importance in bacterial biology. This work demonstrates how spatio-genetically coordinated TPR domain-containing proteins modulate c-di-GMP signaling, contributing to our understanding of biofilm formation in Vibrio species and potentially other bacteria. It also reveals the first evidence of inter-protein interaction via the TPR domains of both partners, challenging the conventional paradigm in which only one bears the domain.
Copyright: © 2025 Mustaree et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

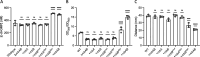



Similar articles
-
Functional analysis of cyclic diguanylate-modulating proteins in Vibrio fischeri.mSystems. 2024 Nov 19;9(11):e0095624. doi: 10.1128/msystems.00956-24. Epub 2024 Oct 22. mSystems. 2024. PMID: 39436151 Free PMC article.
-
Identification of Three New GGDEF and EAL Domain-Containing Proteins Participating in the Scr Surface Colonization Regulatory Network in Vibrio parahaemolyticus.J Bacteriol. 2021 Jan 25;203(4):e00409-20. doi: 10.1128/JB.00409-20. Print 2021 Jan 25. J Bacteriol. 2021. PMID: 33199284 Free PMC article.
-
Calcium signaling controls early stage biofilm formation and dispersal in Vibrio fischeri.J Bacteriol. 2025 Jun 24;207(6):e0007725. doi: 10.1128/jb.00077-25. Epub 2025 May 14. J Bacteriol. 2025. PMID: 40366159 Free PMC article.
-
Implication of environmental factors on the pathogenicity of Vibrio vulnificus: Insights into gene activation and disease outbreak.Microb Pathog. 2025 Jul;204:107591. doi: 10.1016/j.micpath.2025.107591. Epub 2025 Apr 15. Microb Pathog. 2025. PMID: 40246153 Review.
-
Diguanylate Cyclases in Vibrio cholerae: Essential Regulators of Lifestyle Switching.Front Cell Infect Microbiol. 2020 Oct 22;10:582947. doi: 10.3389/fcimb.2020.582947. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194821 Free PMC article. Review.
References
-
- Sampaio A, Silva V, Poeta P, Aonofriesei F. Vibrio spp.: Life Strategies, Ecology, and Risks in a Changing Environment. Diversity. 2022;14(2):97.
-
- Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4(1):1–19. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

